Several public domains were searched for information on the protein products of Ric1 and YLR036c. The information gathered from the websites, along with interpretations are shown below. First, I gathered physical information on the predicted protein products of each gene and scanned the results of a 2 dimensional gel electrophoresis for the products. I then investigated the physical interactions of each protein. The interaction of YLR036c with a recently annotated gene provides strong implications for its function. Many websites were consulted, and those that they failed to retun any information are listed at the end.
Scan for protein products on a 2D gel
I gathered information on the protein products of Ric1 and YLR036c at the PROWL database. The Ric1 protein was predicted to have a molecular weight of roughly 121.6 kDa and an isoletric point of 5.9. The YLR036c was found to have a molecular weight of 23.4 kDa and an isoelectric point of 9.9. A 2d gel generated from S. Cerevisiae proteins (Figure #) was analyzed for proteins appropriate molecular weight and isoelectric point corresponding to Ric1 and YLR036c. No proteins in the region matching the predicted Ric1 protein (coordinates: [5.9,121.6]). Several potential dark spots were observed the expected position of the YLR036c protein (coordinates: [9.9, 23.4]). However, non-linearity in the x-axis at high isoelectric points made identification difficult.
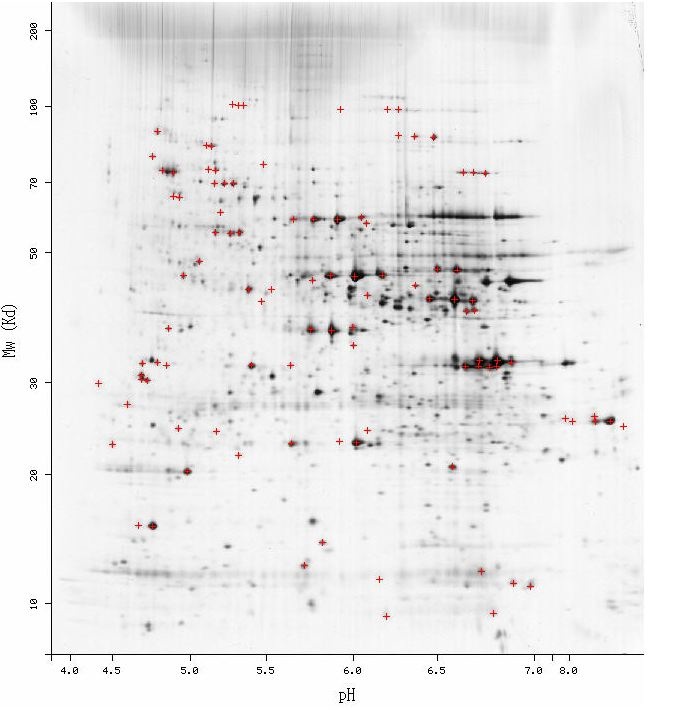
Figure 1 : Two-dimensional gel electrophoresis of proteins from S. cerevisiae cell. A proteins physical properties determine how far it tavels through the gel. Proteins are seperated based on their isoelectric point (x-axis) and molecular weight (y-axis). Dark regions are proteins.
The 2D gel shown above only features the protein found in a S. cerevisiae cell at a single time under a single condition. Since the proteome is constantly changing, 2G gels generated from yeasts at points in the cell cycle and under different conditions will result in distinct 2D gels with different proteins. It is possible that the cell used to produce the 2D gel in Figure 1 did not contain either of the proteins in significant quantities. To overcome the non-constant protein contents of the S. cerevisiae proteome, expression data could be used to find the times when either the Ric1 or YLR036c gene is upregulated. A S. cerevisiae cell could then be lysed during a point in the cell cycle or under condtions that have been associated with the respective gene. Although induction of gene does not always correlate with high concentrations of its protein product, the expression data can act as a guide for finding the optimal conditions for finding the desired proteins. Caution shoulud be exercised in analyzing the 2D gel in Figure 1 for the proteins of Ric1 and YLR036c. The isolectric point and molecular weights that were found at the PROWL database are only predictions and will almost certainly differ from the full length protein. Unknown splicing patterns will likely make the two actual proteins different from the predicted models used to search the 2D gel. The molecular weight and isolectric points of the actual protein products will be necessary before correlating these properties with a 2D gel electrophoresis. |
Ric1: Proteomic information on an annotated gene
Overview: Proteomic information gathered on the interactions of the Ric1 protein agreed with previous research. The MIPS did not report all the high-throughput interactions that were found using the DIP database. The results show the importance of referencing multiple databases for finding data on protein-protein interactions.
Database of Interacting Proteins (DIP)
A visual display of the Ric1 protein's interactions was generated using the Database of Interacting Proteins (DIP) and is provided in Figure 2. Verified physical interactions are found between Ric1, Ypt6p, and Rgp1p, as signified by the green edges connecting their respective nodes. The interactions that connect these three proteins were described in an previous web page and are not discussed here. The two red edges represent tentative interactions between the Ric1 protein and the Lsm2p and Prt1p proteins that are based on high throughput findings.
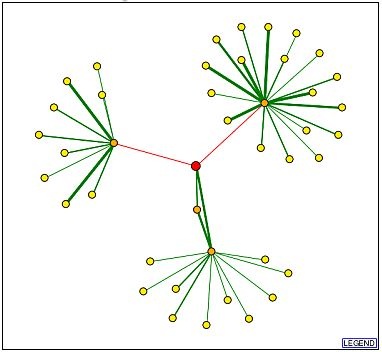
Figure 2: Interactions of Ric1 (red circle). Each circle represents a protein. The lines (edges) either represent a verified protein interaction (green) or a proposed protein interaction (red). The orange circles are 1st shell nodes that have been directly linked to the target (Ric1) protein. All edges extending from the node are drawn. The yellow circles represent 2nd shell proteins, and only the connections between them and the first shell proteins are drawn.
MIPS Database confirms one high-throughput protein-protein interaction
A search of the MIPS Protein-Protein Interaction database was performed. The Ric1 protein was found to engage in a physical interaction with the Lsm2p protein (Figure 3). Interestingly, the the proposed interaction between the Ric1 protein and Prt1p was not found at the MIPS database. A large number of genetic interactions were reported between Ric1 and various genes compared to only the single physical interaction reported in Figure 3 .

Figure 3 : The Ric1 protein (YLR039c) was found to physically interact with the Lsm2p protein using the two hybrid library.
| The physical interaction between Ric1p and Prt1p was listed in the DIP database but not in the MIPS database. This observation reflects the importance of investigating multiple servers for obtaining all available proteomic data rather than trusting in a single database. |
YLR036C: Proteomic information on a non-annotated gene
Overview: An investigation of the YLR036c using proteomic information available from public databases was performed. Data from two web sources were used to find a connection between the YLR036c gene and the recently characterized YKR065c protein. The obtained results indicate that YLR036c localizes to the mitochondria and might be involved in protein import.
Database of Interacting Proteins (DIP)
DIP was searched for physical interactions between YLR036c and other proteins (Figure 4). The database returned that the YLR036c protein interacts with the hypothetical protein YKR065c.
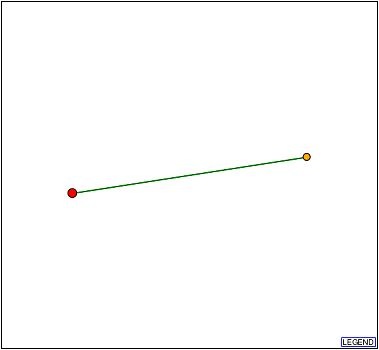
Figure 4: YLR036c protein interacts with the hypothetical protein YKR065c. The green line is evidence that the interaction is supported by a computational method in addition to high-troughput evidence.
MIPS
The MIPS Protein-Protein Interaction database was searched to look for additional physical interactions. The MIPs reported a single physical interaction between YLR036cp and YKR065cp, as verified by individual two hybrid studies (Figure 5). The single interaction agreed with the DIPS graphic.

Figure 5 : Interactions for the YLR036c protein (red) found at the MIPS database. In both experiments, the YLR036c protein functcioned as the prey (or "hit") protein in the interaction.
| The DIPS graphic (Figure 4) features a green edge between the YLR036c protein and the YKR065c proteins. However, it seemed counter-intuitive that non-high-throughput evidence (direct experimental evidence) would exist for the relationship between two non-annotated genes. Searches for direct experimental evidence linking the YLR036c protein and the YKR065c protein yielded nothing. Assuming that no study has been conducted, the fact that two high-throughput experiments have individually observed a physical interaction between the YLR036c and YKR065c proteins is the most likely justification for using the green edge in the DIPS graphic. |
SGD: YKR065c's secret identity
I searched the SGD database for information of the YKR065c protein that interacts with YLR036cp. YKR065cp (Pam17) was recently identified as the sixth motor subunit of the presequence translocase-associated motor (PAM) in the mitochondrial matrix (van Der Laan et al. 2005). PAM functions in the import of proteins into the mitochondria. Although Pam17 mutants have impaired PAM function, they are still viable (Giaever G et al. 2002). Within the PAM unit, Pam17 contributes to the formation of the Pam16-Pam18 complex, which is necessary for the regulation of mtHsp70, the preprotein-binding matrix heat shock protein 70. Pam17 is anchored to the inner membrane of the mitochondria, and its molecular function remains unknown.
The linkage of YLR036c to Pam17 (YKR065c) is a major step in its characterization.
The discovery of a physical interaction between YLR036cp and Pam17p is a best case scenario. Assuming that the interaction is valid, we learn much from the connection. First, Pam17p is specific to the inner membrane of the mitochondrion. This suggests that the YLR036c protein product is also found in the inner membrane region of the mitochondrion. Second, Pam17 has been identified as a member of the presequence translocase-associated motor through mutation studies (van Der Laan et al. 2005). Also, it is specifically known to interact with the Pam16-Pam18 complex. The knowledge of the interactions of the Pam17 protein will allow inferences and predictions to be made about the YLR036c and to guide further testing. For more on future experiments, see the final segment of this web page. The case of YLR036c and Pam17p highlights the usefulness of high throughput methods in characterizing genes. The physical interaction between YLR036c and YKR065c (Pam17) was identified in 2000, when no information was available on either gene (Ito T et al.). The information now known about Pam17 (YKR065c), however, can now be applied to the determination of the YLR036c gene. The collection of the high-throughput data is essentially providing researchers with leads that will fasten the rate of genetic discovery. From this identification, I predict that the YLR036c protein will be a protein that localizes to the mitochondria and functions in the presequence trarnslocase-associated motor. This hypothesis is entirely different than predictions made in previous web assignment, which were based on much weaker evidence. Also, protein interaction data should supersede the expression data since analysis of proteins is more directly associated with a gene's function. |
Searches in the following databases did not return any information for either gene: Protein Data Bank (PDB), SWISS-2DPAGE, YRC Two-Hybrid Analysis, and the Enzyme and Metabolic Pathways (EMP). In addition, both genes were absent in PDF data reported by Benno et al.
Future experiments should build off the interaction between YLR036c and Pam17 The interaction between the YLR036c the Pam17 proteins is a major step towards identification of the proteins function. The first step I wold propose is performing an immunoprecipitation reaction to conclusively determine that YLR036cp interacts with Pam17p. Assuming that the interaction is verified by the immunoprecipitation, I would next prepare a monoclonal antibody against the YLR036c protein and perform an immunostaining experiment. If the YLR036c protein is not obsered on a membrane or in the cytoplasm, its interaction with Pam17 will allow us to investigate the mitochondria. Yeast two-hybrid studies, or immunoprecipitation, should be performed between the YLR036c protein and other proteins on the presequence translocase-associated motor unit, which are involved with Pam17. I also propose a mutation study for YLR036c that will analyze specifically for loss of protein import into the mitochondria. More generally, function in the mitochondria should be observed. The procedure should resemble the mutation study conducted by van der Laan et al. in determining the function of Pam17. A mutation study might also determine a function unrelated to the PAM complex in the mitochondria. If the studies come up negative, high-throughput methods such as the yeast-two hybrid method will remain the mosteffecient methods for providing new insights for future experiments and possible functions. |
References
Giaever G, et al. (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418(6896):387-91.
Ito T, et al. (2000) Toward a protein-protein interaction map of the budding yeast: A comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc Natl Acad Sci USA 97(3):1143-7.
van der Laan M, et al. (2005) Pam17 Is Required for Architecture and Translocation Activity of the Mitochondrial Protein Import Motor Mol Cell Biol 25(17):7449-7458.
Ben Whigham's Genomics Home Site.
Genomics Course Web Site.
© Copyright 2005 Department of Biology, Davidson College,Davidson, NC 28036
Send comments, questions, and suggestions to: bewhigham@davidson.edu