From the cDNA sequences found in Genbank for Mus musculus, Drosophilia melanogaster, C. elegans, Arabidopsis thaliana, S. cerevisiae and Homo sapiens, we can use MacDNAsis to determine the largest open reading frame (ORF) and determine the proteins molecular weight by translating it.
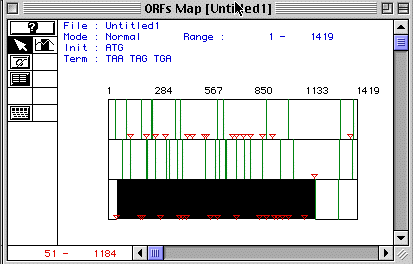
Figure 1. ORF Map of human actin

Figure 1 shows a display of human actin's ORF Map. Each row demonstrates the ORF map for each reading frome. The third frame contains the lagest ORF beginning at the start codon, represented by the red triangle, and ending at the stop codon, represented by the green line. This ORF lasts from nucleotide 51-1184. Analysis using MacDNAsis predicts this ORF of the protein to have a molecular weight of 42048.81 Daltons.
Figure 2. Kyle and Doolittle hydrophobicity plot
for human actin
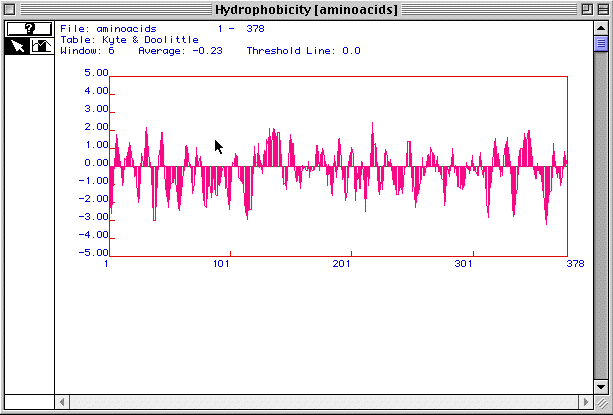
Figure 2 is a hydrophobicity plot for actin generated by MacDNAsis.
Peaks above 2.00 suggest that a protein is an integral membrane protein.
With at least 3 peaks above 2.00, actin is most likely an integral membrane
protein.
Figure 3. Hopp and Woods antigenicity plot
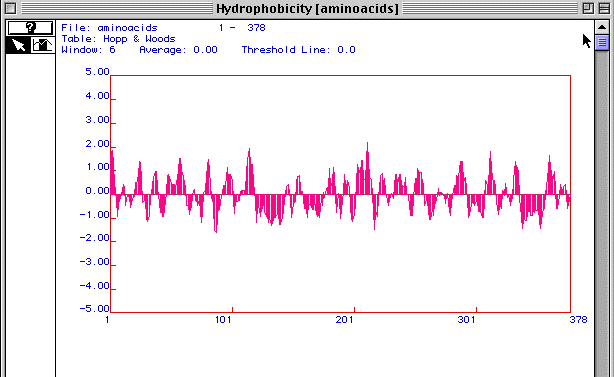
Figure 3 is an antigenicity plot for actin generated using MacDNAsis. It shows regions of hydrophilicity (disregard the hydropathy title in the figure). Those regions that are most positive are most likely the most hydrophillic. Therefore these peptide regions would be the ones that exist outside the cell membrane. These regions would be the most likely candidates to target as epitopes for developing monoclonal antibodies to actin.
Figure 4. Secondary Structure of Actin
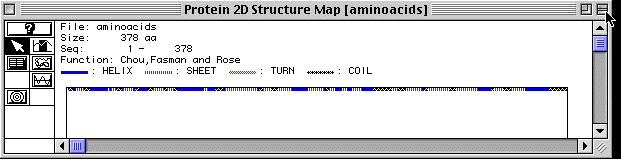
Figure 4 shows the secondary structure of actin as analyzed by MacDNAsis. There are regions of alph helices, beta-pleated sheets and coils; however actin, apparently is a straight protein because no turns are shown by the MacDNAsis analysis. A RasMol image of actin is provided to further visualize the protein. (The image is of actin-binding protein because no actin image was available)
Figure 5. Amino Acid Conservation Comparison

Figure 5 compares the amino acid residues 301-500 for all 6 species.
The figure uses the single-letter abbreviation to denote amino acids.
Highlighted letters show amino acids that are conserved between all the
species. This figure suggests that actin is well conserved and may
not have changed much during the course of evolution. This also indicates
that actin is a very primitive protein since it is found both in yeast
and humans in very similar forms.
Figure 6. Phylogenetic Tree
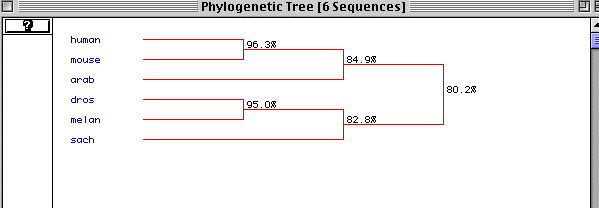
Figure 6 is the predicted phylogenetic tree generated by MacDNAsis.
The different species are ordered with the most primitive at the bottom.
It shows that actin is very well conserved with an 80.2% conservation between
humans and yeast. Normally a conservation of 20% is deemed significant.
This figure supports the assumptions made from figure 5 that actin is well
conserved and very primitive.