This web page was produced as an assignment for an undergraduate course at Davidson College.
My Favorite Yeast Proteins:
TOP1 and YOL008W
TOP1 and YOL008W are Saccharomyces cerevisiae genes that are located on chromosome XV. TOP1 (YOL006C) encodes Topoisomerase 1 (an enzyme that cleaves single-stranded DNA) and plays a role in important cellular functions such mitotic recombination, DNA elongation, transcription regulation, and mitotic chromosome condensation. YOL006C is a non-annotated gene, which means there is very little known about its protein's function. With the help of online genomic databases, I have already characterized TOP1 and YOl008W through sequence analysis and expression patterns. In this assignment, I will use online proteomics databases to learn more about the functions of the TOP1 and YOL008W proteins. While sequence analysis and expression patterns can be very helpful to predict the role of proteins, neither method of analysis can represent the role of a protein more accurately than the analysis of the protein itself and its interactions in the cell.
To view my web page on the sequence analysis of these two genes, go to My Favorite Yeast Genes: TOP1 and YOL008W.
To view my web page on the expression pattern analysis of TOP1 and YOL008W, go to My Favorite Yeast Expression: TOP1 and YOL008W.
TOP1 (YOL006c)
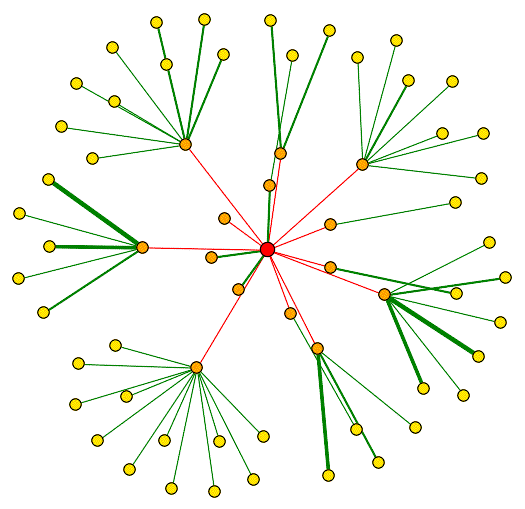
I searched DIP to find the proteins with which Top1 interacts. Figure 1 shows Top1 interacting with 14 different proteins (orange nodes), and for many of these proteins in the first shell, the diagram depicts them interacting with other proteins (yellow nodes). Several of the proteins it is shown to interact with have unknown functions, but the majority of the proteins that TOP1 interacts with are involved some DNA process, such as topological change, transcription regulation, or chromatin assembly. This makes sense that Top1 interacts with proteins involved in these type of functions because Top1 is involved in many of these processes and functions to cleave single-stranded DNA. A list of the proteins that Top1 interacts with in Figure 1 is shown in Figure 2.

Figure 1. Screen shot of protein interactions for Top1. The red dot,
or node, in the center represents Top1. The orange dots represent
the first shell of nodes, or the proteins the Top1 directly interacts with.
The yellow dots represent
the second shell of nodes. The nodes are connected by edges (lines) and a thicker
edge width indicates greater reliability for the interaction. Image from DIP,
2003 <http://dip.doe-mbi.ucla.edu/dip/DIPview.cgi?PK=1705>.
Gene |
Protein Function or Process |
| TOP1 | involved in DNA topological change |
| RPC82 | DNA-directed RNA polymerase activity, involved in transcription from Pol III promoter |
| RIF2 | telomeric binding; telomeric maintenance |
| CLU1 | translational initiation, mitochondria organization and biogenesis |
| YFR011C | function unknown; located in cytoplasm |
| RPD3 | histone deacetylase; involved in chromatin silencing, mitotic recombination, DNA-dependent transcription regulation |
| OCA1 | tyrosine phosphatase activity; responds to oxidative stress |
| REB1 | regulation of transcription, transcription termination |
| HTB2 | chromatic assembly/diassembly |
| TOF2 | DNA topological change |
| YMR233W | unknown function |
| HMRA2 | unknown function |
| ACO1 | citrate and proprionate metabolism, glutamate biosynthesis |
| SPT16 | transcription regulation and elongation |
Figure 2. Descriptions of proteins that interact with Top1. This is a list of proteins from Figure 1 that interact with Top1. These proteins are represented by the orange nodes. The first protein in the list is the orange node at "12:00" in Figure 1, and the list continues in a clockwise fashion. The brief descriptions of each protein were taken from SGD, 2003.
Next I looked at a PDF file called "Benno Fgiure 1" from a paper by Stan Fields, Peter Uetz, and Benno Schwikowski. The file shows 2,709 interactions between 2,039 different proteins (a very large file). I searched for TOP1 to see which proteins it interacted with. Figure 3 shows TOP1 with its protein interactions. The figure shows TOP1 interacts with TOF2, TOF1, and YMR233W. Top1 was shown to interact with all of these proteins from the DIP database (Figure 1), so the data from these two sources are consistent with one another. TOP1 is connected to the proteins by black lines, which means that they have different cellular roles and localizations, so it is interesting that all of these proteins interact given their different functions. The yellow boxes indicate unknown roles for the proteins.
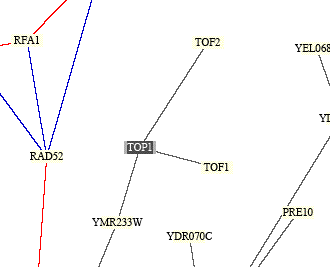
Figure 3. TOP1 interactions from "Benno Figure 1" by Fields, Uetz, and Schwikowski. The proteins are shown in boxes and the lines indicate between which proteins interactions occur. TOP1 is shown to interact with three proteins. The function of these proteins are listed in Figure 2. The black lines mean the proteins have different cellular roles and different localizations and the yellow boxes mean unknown roles of the proteins. Image from <http://occawlonline.pearsoned.com/bookbind/pubbooks/bc_mcampbell_genomics_1/medialib/seq/Benno/NB_Figure1color.pdf>.
Figure 4 shows another protein interaction image for TOP1 from the Pathcalling Yeast Interactive Database. In this figure, TOP1 directly interacts with RPA14, RPA34, YMR233W, and TOF1. From the previous figures, TOP1 was shown to interact with YMR233W and TOF1, but the RPA14 and RPA34 are new interactions. From this database, RPA34 is listed as RNA polymerase 1 subunit and RPA14 is RNA polymerase 1 subunit A14, so both of these proteins have roles in transcription. It makes sense that Top1 would interact with other proteins that have roles in transcription because Top1 also functions during transcription.
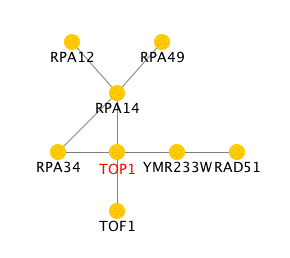
Figure 4. Protein interactions for TOP1. Top1 is located in the center (red) and the lines show its interactions with other proteins. Image from Pathcalling Top1 Information Page, 2002 <http://portal.curagen.com/extpc/com.curagen.portal.servlet.PortalYeastGene?geneIdIn=5169>. (Image generated with information from "Uetz, et al., Nature 403, 623-627 (2000)".)
The MIPS database also contains functional information for Top1. Figure 5 shows a list of the proteins that Top1 was found to interact with (protein-protein interactions). The results come from several different papers, and many of the interactions were determined by the yeast two-hybrid interactions The yeast two-hybrid system is a high-throughput method that determines interacting proteins by the transcription of a reporter gene. MIPS contains information on 13 different interactions for Top1. Some of the proteins that Top1 is shown to interact with are consistent with the data from previous databases, such as RPA14, RPA34, YMR233w and Tof1, but some of the proteins are new interactions that I have not previously found (such as Trf4, Ynl099cp, and Sgs1). The proteins that have not been mentioned previously have roles in genomic stability, ATPase activity in the presence of DNA, cell differentiation, cell cycle and DNA processing, organization of chromosomes. Some of these proteins had unknown functions, and many of the proteins were involved in the cell cycle and DNA processing. The only process that I was slightly surprised about initially was that Tof1 interacts with a protein involved in cell differentiation. But when cells differentiate, many new genes are turned on and transcribed, so it would make sense that Top1 (an enzyme that cleaves single-stranded DNA) would interact with a protein involved in this process. Interestingly, the localization of these proteins included the nucleus, the cytoplasm, and mitochondria. MIPS lists Top1 as being localized in the nucleus, yet it interacts with all of the above proteins. Perhaps Top1 does not always remain in the nucleus, or perhaps some of the other proteins are not permanently localized in the cytoplasm. In addition, the MIPS database listed that Top1 was found to interact in complexes of proteins that were involved in transcription, DNA maintenance, and chromatin structure, which is what we have just observed.
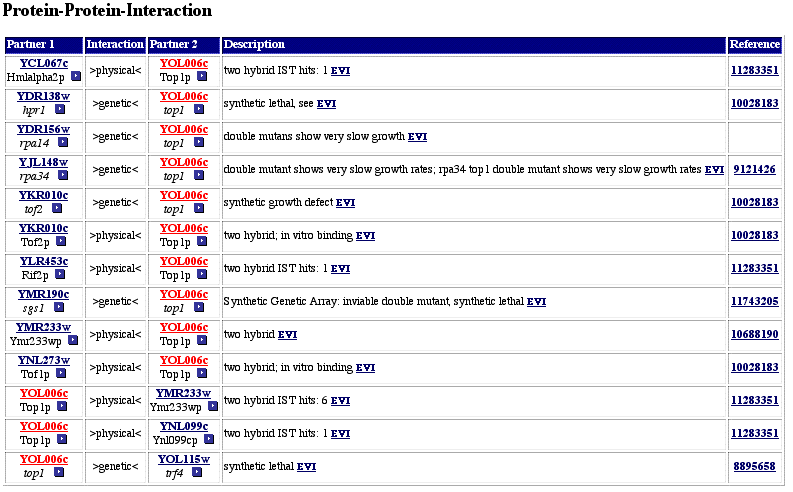
Figure 5. Protein-protein interactions for Top1 (Yol006c). This figure shows the proteins that Top1 was found to interact with, primarily from the yeast two-hybrid method. Top1 (Yol006c) is shown in red. Image from MIPS, 2003; http://mips.gsf.de/genre/proj/yeast/searchEntryAction.do?text=YOL006c.
Some of the databases I searched did not contain any information on Top1 and some contained sequence information but did not contain any functional information about the protein. Below is a list of databases that did not yield any new information.
Enzymes and Metabolic Pathways: no information on Top1.
Interaction Sequence Tags: no information on Top1.
PDB: contains structural information, but no functional information.
PROWL: contains sequence information, but no functional information.
2D Database: The molecular weight and the isoelectric point of the protein are needed to determine the protein's theoretical location on the gel. The molecular weight for Top1 is 89.9 kD and its isoelectric point is 9.38. However, I was unable to find any information for Top1 on the gel.
KEGG: no information found.
Y2H: This database listed Top1 as interacting with YMR233w. This information is consistent with information from other databases, but it provides no new information.
TRIPLES: There was an entry listed for YOL006c, but none of the information was very helpful in determining the function of Top1. The only information I could extract from this database was when a transposon was inserted at codon 311 (clone ID V22A7). All of the phenotypic data was wildtype except for two different assays. This mutant showed mild sensitivity to growth media of hygromycin, indicating that the gene potentially functions in the cell wall or biosynthesis. Top1 is not known to function in the cell wall, but could possibly function in biosynthesis. It also showed mild sensitivity to EGTA, indicating that this mutant is cation sensitive.
ExPaSy: This database only led me to sequence information for Top1.
WIT?: no information found.
Final Thoughts and New Experiments
After analyzing the sequence, expression patterns, and protein functions and interactions for Top1, I have found information to support the ideas that Top1 is an enzyme that functions to cleaves single-stranded DNA in cellular processes such as mitotic recombination, DNA elongation, chromatic assembly/disassembly, transcription regulation, and mitotic chromosome condensation. However, some of the proteins it interacts with are involved in slightly different cellular processes (such as cell differentiation) and are located in places other than the nucleus. I have gathered data that supports the previous known roles of DNA Top1, but I have also expanded its cellular roles. We know some information about Top1, but it is most likely that we have more to learn about all of its protein interactions. Some databases listed above contained protein interaction information, but the majority of the databases did not. The field of proteomics walks a fine line between what we already know and what we do not yet understand. The next logical step from here would be to design experiments from which we can better understand the function of Top1 in the cell.
Localization of Top1
Most databases say that Top1 is located in the nucleus of the cell, but I thought it was very interesting that it interacts with proteins that are in the cytoplasm and mitochondria. I would like to determine if Top1 is located in other places in the cell other than the nucleus. To do this, I would use immunofluorescence to determine the localization of the protein. I would use an antibody that is specific to Top1 and covalently link it to fluorescent dye. I would use yeast cells that are in different stages of the cell cycle, make the membranes permeable with detergent, and fix them to them to slides. Then I would put the antibodies over the slide so they had a chance to bind with Top1, and then view the fluorescence under a microscope. I would expect to see a lot of fluorescence in the nucleus because Top1 is located there. I would also expect to see some fluorescence in the mitochondria (since the mitochondria have its own DNA, I would expect that Top1 would also be needed in that organelle) and perhaps some fluorescence would show up in the cytoplasm as well. It would be very interesting to find Top1 in the cytoplasm because this could mean that it is able to travel back and forth, or perhaps it has an entirely new function that we are unaware of yet. If there is no Top1 seen in the cytoplasm, then perhaps the mitochondria has its own Top1 separate from that of the nucleus (or maybe the method was not sensitive enough to detect it). This experiment could also help to determine at what time points during the cell cycle Top1 is expressed and in what amounts. Cells can be observed at different time points during the cell cycle and the relative level of fluorescence would give an indication of the amount of Top1 that is present. I would expect that Top1 would be highly expressed during replication.
YOL008W
I searched DIP to find information on the proteins with which YOL008W interacts (Figure 6). This search did not yield as many results as it did for TOP1, but this is to be expected since the function of YOL008W is unknown. The primary protein with which YOL008W interacts is ATP14 (orange node in Figure 6). ATP14 has hydrogen-transporting ATP synthase activity, it is involved in the process of ATP synthesis coupled proton transport, and its cellular component is proton-transporting ATP synthase complex (SGD, 2003 <http://genome-www4.stanford.edu/cgi-bin/SGD/locus.pl?locus=ATP14>). According to MIPS, ATP14 is localized in the mitochondria. Previously, I predicted that YOL008w was localized in the cytoplasm or nucleus, but this data presents a new angle. Perhaps YOL008w can also be localized in the mitochondria, or perhaps ATP14 can move back and forth between the mitochondria and the cytoplasm. These data expand the previous ideas of localization for YOL008w.
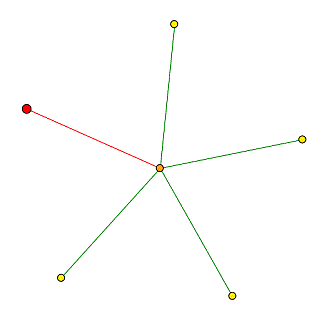
Figure 6. Protein interactions for YOL008w. The red node represents YOL008w, and it is known to interact with only one other protein. Image from DIP, 2003 <http://dip.doe-mbi.ucla.edu/dip/DIPview.cgi?PK=4689>.
In Figure 6, ATP14 is connected to four other nodes; the functions of these proteins are described in Figure 7. Two of these proteins show ATPase activity, one's function is unknown, and the fourth gene functions in biosynthesis. Knowing that YOL008w interacts with an ATP synthase, and this ATP synthase interacts with other ATP sythase proteins, perhaps YOL008w has ATP synthase activity as well. It is also possible that it has a role in transport or some type of biosynthesis. These findings are somewhat consistent with what I deduced from sequence analysis and expression patterns, as I predicted that it may have a role in biosynthesis or transport from ER to Golgi. However, these findings also expand the range of possibilities for the role of YOL008w. Of the four proteins that interact with ATP14, some are localized in the cytoplasm and some are localized in the mitochondria.
Gene |
Protein Function or Process |
| AYR1 | phosphatidic acid biosynthesis; located in cytoplasm, ER, and lipid particles |
| YIR036C | function unknown, located in cytoplasm |
| ATP1 | hydrogen transport ATP synthase activity; ATP synthesis coupled proton transport |
| ATP2 | hydrogen transport ATP synthase activity; ATP synthesis coupled proton transport and age dependent metabolic decline |
Figure 7. Descriptions of proteins that interact with ATP14. These proteins represent the yellow nodes in Figure 8 and are the secondary shell nodes for YOL008W. The protein descriptions are from SGD, 2003.
Next I searched the Pathcalling Yeast Interactive Database for more information about the protein interactions. YOL008w was not shown to interact with any proteins (Figure 8), illustrating the fact that very little is known about this ORF.

Figure 8. Protein interactions for YOL008W. The Pathcalling database showed no known interactions for YOL008W. Image from Pathcalling YOL008w Information Page, 2002 <http://portal.curagen.com/extpc/com.curagen.portal.servlet.PortalYeastGene?geneIdIn=5171&keywordIn=YOL008w>. (Image generated with information from "Uetz, et al., Nature 403, 623-627 (2000)".)
The MIPS database cotained only one protein-protein interaction for YOL008w (Figure 9). This screen shot shows YOL008w was found to interact with Atp14 in a yeast two-hybrid experiment. According to MIPS, Atp14 is located in the mitochondria, functions in the oxidative phosphorolation pathway and is involved in ion transport. This data is consisten with the data from DIP, but again, it is interesting that ATP14 is localized in the mitochondria because MIPS lists YOL008w as being in the cytoplasm (along with my own prediction). There was no information given for the complex interactions.

Figure 9. Protein-protein interactions for YOL008w. Image from MIPS, 2003; http://mips.gsf.de/genre/proj/yeast/searchEntryAction.do?text=YOL008w.
Many databases that I searched did not contain any information on YOL008w. This is to be expected since YOL008w is an unannotated ORF, but it is somewhat frustrating when you want to know more about it! Below is a list of searches that did not yield any results.
Schwikoski Figures: no available information for YOL008w.
Enzymes and Metabolic Pathways: no information for YOL008w.
2D Database: no information for YOL008w.
Interaction Sequence Tags: no information for YOL008w.
KEGG: no information for YOL008w.
PDB: no information for YOL008w.
Y2H: no information for YOL008w.
PROWL: contained sequence information, but no functional information for YOL008w.
TRIPLES: no information for YOL008w.
ExPaSy: This database only led me to sequence information for YOL008w (no functional information).
WIT?: no information found.
Final Thoughts and Future Experiments
After analyzing the sequence, expression patterns and protein interactions for the unannotated ORF YOL008w, we have come a little closer to elucidating its role. However, many databases resulted in dead ends, and the information I have gathered is only my best prediction; nothing is for certain. The information from the protein interaction databases expanded my previous predictions for this ORF so that now, it may be located in the cytoplasm or nucleus, but it is also possible that it functions in the mitochondria at times. It interacts with ATP synthase, but it also could have a role in lipid metabolism, biosynthesis or transport. The data from the expression patterns indicated that it may be involved in trasport and the data from the protein interactions suggests that it may have ATP synthase activity, so these two predictions could be consistent with one another depending on its actual function. Regardless, all of this information basically shows that we do not know the role of YOL008w, but based on our predictions, we can develop experiments to determine if our predictions have any truth.
Localization of YOL008w?
Since the location of YOL008w is still unknown, I think it would be very helpful to determine where in the cell this protein is located. This experiment would be very similar to the experiment for determining the localization of Top1. I would use immunofluorescence to find out where YOL008w is located and also to determine at which point in the cell cycle the protein is present. An antibody that is specific to YOL008w would be labeled with immunofluorescent dye, and then the location of the dye would be detected within the cell. Since YOL008w is known to interact with ATP14, a protein from the mitochondria, I would predict to see immunofluorescence in the mitochondria. I also predicted that it was localized in the cytoplasm, so it would be interesting to see if it is found in the cytoplasm as well.
Protein Interactions?
There are very few known protein interactions for YOL008w, but I have a feeling it interacts with more proteins than only ATP14. I would like to conduct a specific experiment to find out more about the proteins that YOL008w interacts with. I would use the Yeast Two-Hybrid (Y2H) method to find out more about its protein interactions. The YOL008w protein would be used as "bait" and attached to a binding domain of a transcription factor. Then, "prey" proteins would be fused to the activation domain of a transcription factor. A different prey protein would be placed in every cell with the bait protein. If the YOL008w interacts with any of the different prey proteins, RNA polymerase will be triggered to transcribe the gene. The gene that is transcribed would be His3 (produces histidine). Any cells in which the bait and prey protein interact will not produce histidine and therefore will not be able to survive (Campbell, 2003). So, any colonies of surviving yeast cells means that YOL008w interacts with the prey protein. First I would test to see if YOL008w does in fact interact with ATP14. I would also test different ATP synthases to see if YOL008w interacts with any of those proteins. If I found that YOL008w was specificallly localized in the mitochondria (from the previous experiment), then I might use all of the proteins in the mitochondria as prey proteins. I also would test proteins from other parts of the cell (such as the cytoplasm) to get a more complete view of which proteins YOL008w interacts with.
Vital protein?
I would also like to know if YOL008w is required for the cells to grow, or if a mutation or knockout in this gene would cause the cell to die. For this experiment, I would mutate the gene in some yeast cells and delete it in others to see if the cell would survive. If the cell dies when the gene is mutated, then this mutation is a lethal mutation and a functional copy of this gene is necessary for yeast survival. If the cell dies when the gene is deleted but is able to survive when the gene is mutated, then perhaps the cell can survive with a less functional copy (or maybe the mutation doesn't affect its function at all). If the cell survives when the gene is deleted, then it is not required for the yeast cell to survive, or at least not to survive in optimum conditions. From here, I would exposed the cells with the knocked out gene to different environmental conditions, such as high salt salinity, different temperatures, and different growth medias and look for any difference in the yeast's phenotype or its ability to survive.
References
Campbell, A. Malcolm and L. J. Heyer. 2003. Discovering Genomics, Proteomics, and Bioinformatics. Benjamin Cummings: San Francisco.
[DIP] Database of Interacting Proteins. 2003. <http://dip.doe-mbi.ucla.edu/dip/Search.cgi?SM=3>. Accessed 2003 Nov. 11.
PathCalling Yeast Interaction Database. 2002. <http://portal.curagen.com/extpc/com.curagen.portal.servlet.PortalYeastList?modeIn=List>. Accessed 2003 Nov. 16.
[SGD] Saccharomyces Genome Database. 2003. <http://www.yeastgenome.org/>. Accessed 2003 Nov. 11.
MIPS Comprehensive Yeast Genome Database. 2003. <http://mips.gsf.de/genre/proj/yeast/index.jsp>. Accessed 2003 Nov. 19.
Back to Sarah's Homepage for Genomics, Proteomics and Bioinformatics.
______________________________________________________________________________
Davidson College Biology Department
Davidson College Genomics Homepage
Sarah's Molecular Biology Homepage
Please send questions and comments to Sarah Baxter at sabaxter@davidson.edu. Thanks!