*This web page was produced as an assignment for an undergraduate course at Davidson College*
(back)
On previous webpages, I have captured and analyzed data on the sequences and expression profiles of my favorite yeast genes, Vas1 and YGR093w. On my final webpage, I will explore data concerning the structure and interactions of Vas1 and YGR093ws' protein products. A number of databases contain information about what proteins interact with each other, as well as functional and locational predictions based on amino acid structure. I will present the most important available data and attempt to synthesize this information. Afterwards, I will compare these findings to my previous hypotheses about these genes based on their sequence and expression.
As discussed on my favorite yeast gene webpage, Vas1 (valyl-tRNA synthetase) catalyzes the binding of valine and an appropriate tRNA molecule to form valyl-tRNA. Vas1 is a member of a group of genes known as aminoacyl-tRNA synthetases. Each of these genes catalyzes the formation of a specific aminoacyl-tRNA molecule. This process is integral to protein translation. Without the action of Vas1, valine could not be incorporated into any proteins that yeast synthesized. Vas1 functions both in the cytoplasm of yeast as well as the mitochondria. A glimpse at the proteins that Vas1 has been found to interact with will simultaneously show the utility and current limitations of proteomics databases.

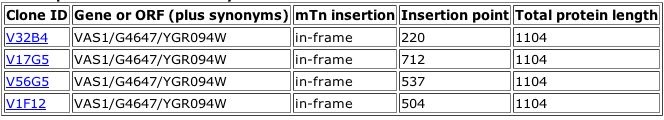
Figure 1. Results of a TRIPLES database search for the phenotypes of mutants with insertions within the Vas1 gene. These four "clones" each represent a population of yeast with a verified transposon insertion in this gene. From the information above, it is clear that each insertion left the original gene in the correct reading frame. More detailed information is available about each clone. Data on one Vas1 mutant is shown below.
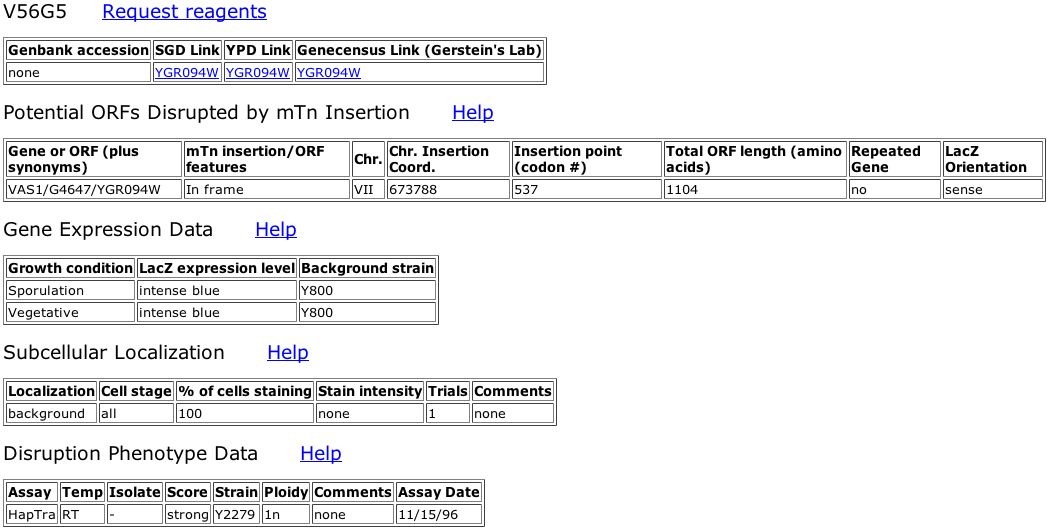
Figure 2. Information about TRIPLES database Vas1 mutant, V56G5. The LacZ transposon insertion occurred at bp 673788 on Yeast chromosome VII. This location corresponds to the 537th codon of Vas1, very near the middle of final 1104 amino acid polypeptide. The LacZ gene inserted in the correct orientation ("sense"). Upon analysis, the expression level of this mutated Vas1 gene seems constitutively high since LacZ stained intense blue during sporulative and vegetative states. LacZ, and correspondingly Vas1, was not localized at any specific subcellular location within the yeast cells (background). Like the other three Vas1 mutants in the TRIPLES database, V56G5 showed a score of strong when assayed for inviability as a haploid.

Figure 3. The score table for TRIPLES database Disruption Phenotype Data. While potentially subjective, this scoring rubric provides an idea of the strength of observed growth differences between wild-type and mutant clones of yeast.
As stated above, the four Vas1 mutants in the TRIPLES database all were tested for haploid inviability. All mutants showed strong differences in growth between mutant and wildtype. Since Vas1 is essential to production of any polypeptide with the amino acid valine, it seems reasonable that a disruption in this gene would have major consequences in numerous ways affecting growth.
The only differences between these four clones' LacZ expression patterns were their cellular components, or location. V56G5 did not show any localization of LacZ expression. While clones V32B4 and V17G5 expressed LacZ in their mitochondria, V1F12 showed cytoplasmic LacZ localization. The insertion of an entire gene into another could very easily alter the original gene's cellular component. Thus, the results of V56G5 are not surprising. When expression was localized, it occurred in either the mitochondria or the cytoplasm. This corresponds to published information about Vas1, in which researchers showed that Vas1 is alternatively spliced into two forms, one functioning in the mitochondria and the other in the cytoplasm of yeast (Chatton et al., 1987).
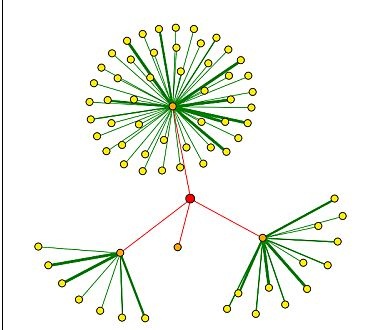
Figure 4. Results of a DIP search for proteins shown to bind Vas1. The red dot (node) represents Vas1. Red lines connect Vas1 to orange nodes, which are proteins shown to directly interact with the Vas1 protein. Yellow nodes represent proteins that interact secondarily with Vas1 through the corresponding orange nodes. Thicker lines correspond to more experiments that support these interactions. Clockwise from the top, Vas1 protein directly interacts with calmodulin (CMD1) , TAF6, Ribonuclease H (RNH1), and CDC14.
| Gene Ontology Term | CMD1 | TAF6 | RNH1 | CDC14 |
| Biological Process | -budding -cytoskeleton organization and biogenesis -mitosis |
-establishment and/or maintenance of chromatin architecture -transcription initiation from -Pol II promoter -histone acetylation -protein amino acid acetylation -chromatin modification -G1-specific transcription in mitotic cell cycle |
-DNA replication -cell wall organization and biogenesis |
-protein amino acid dephosphorylation -regulation of exit from mitosis |
| Molecular Function | -calcium ion binding | -general RNA polymerase II transcription factor activity | -Ribonuclease H activity | -phosphoprotein phosphatase activity |
| Cellular Component | -bud neck -incipient bud site -central plaque of spindle pole body -shmoo tip -cytoplasm -bud tip |
-transcription factor TFIID complex -SAGA complex |
-cell | -RENT complex -spindle pole body -nucleolus -nucleus |
Table 1. Basic Gene Ontology data (from the DIP node database) about the four "1st shell nodes" identified by DIP.
DIP shows the Vas1 protein interacting directly with four varying genes. While TAF6 and CDC14 both function in protein protein (by acetylation and dephosphorylation, respectively), CMD and RNH are both involved in mitosis and DNA replication. The information from this database highlights the limited nature of available data. While it could be explained that Vas1 interacts with these diverse proteins because protein translation is a ubiquitous process, it would be extremely difficult to predict the function of Vas1 solely from its interactions with these proteins.
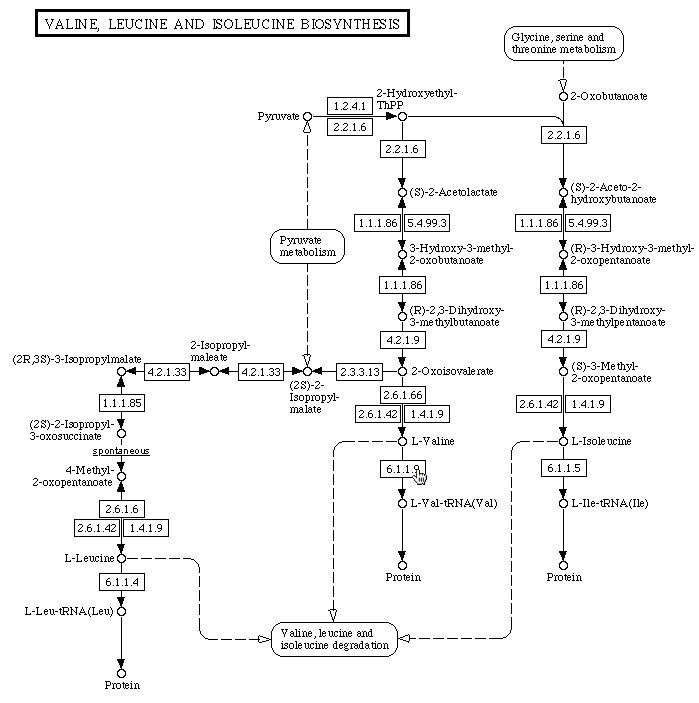
Figure 5. Depiction from the KEGG Pathway website of the metabolic pathway of Valine, Leucine and Isoleucine biosynthesis. Named molecules are connected by arrows, each depicting a chemical reaction. The numbers on each arrow correspond to the metabolic enzyme(s) that catalyzes each reaction. The cursor points to 6.1.1.9, or Vas1, which catalyzes the formation of L-Valine-tRNA molecules by ligating L-Valine to the appropriate amino acid.
ExPASy Biochemical Pathways - Found information very similar to that on KEGG Pathway website.
Protein Data Bank - No available on Vas1 protein.
PROWL - All links to Vas1 and Vas1 protein data are broken.
Y2H Database - No data with Vas1 as bait or prey protein.
Searching through the aforementioned protein databases, I found no information contradicting previous published data about Vas1. On my favorite gene webpage I established that this gene codes for an enzyme vital for the incorporation of valine in polypeptides. With that in mind, results from the TRIPLES database clones with insertions in the Vas1 chromosome region reinforced the importance of this gene. Disrupting the formation of functional Vas1 protein strongly affected growth in mutants. Without the catalyzing action of Vas1 protein, valyl-tRNA is not produced at a sufficient rate. This affects all protein production, and thus affects growth. While very intriguing in theory, the available protein interaction data did not illuminate the function of Vas1p. Without prior knowledge of Vas1's biological process, it would be impossible to determine from the results of DIP and TRIPLES database searches. Next, I will use these databases to refine my hypotheses about the function of the non-annotated gene, YGR093w.
On my favorite yeast genes webpage, I analyzed the results of a search for conserved domains based on the predicted amino acid sequence of YGR093w.

Figure 6. The Conserved Domain website provides information on protein domains that align well to the provided amino acid sequence. YGR093W aligns very well to the proteins CwfJ_C_1 (E(= 2e^-35) and CwfJ_C_2 (E=7e^-24), where E = the probability of two unrelated domains randomly aligning as or more closely.
The CwfJ family proteins are involved in the mRNA splicing complex. I previously hypothesized that YGR093w might also function somehow in mRNA splicing because of its very close alignment to the N-terminus of these two proteins.
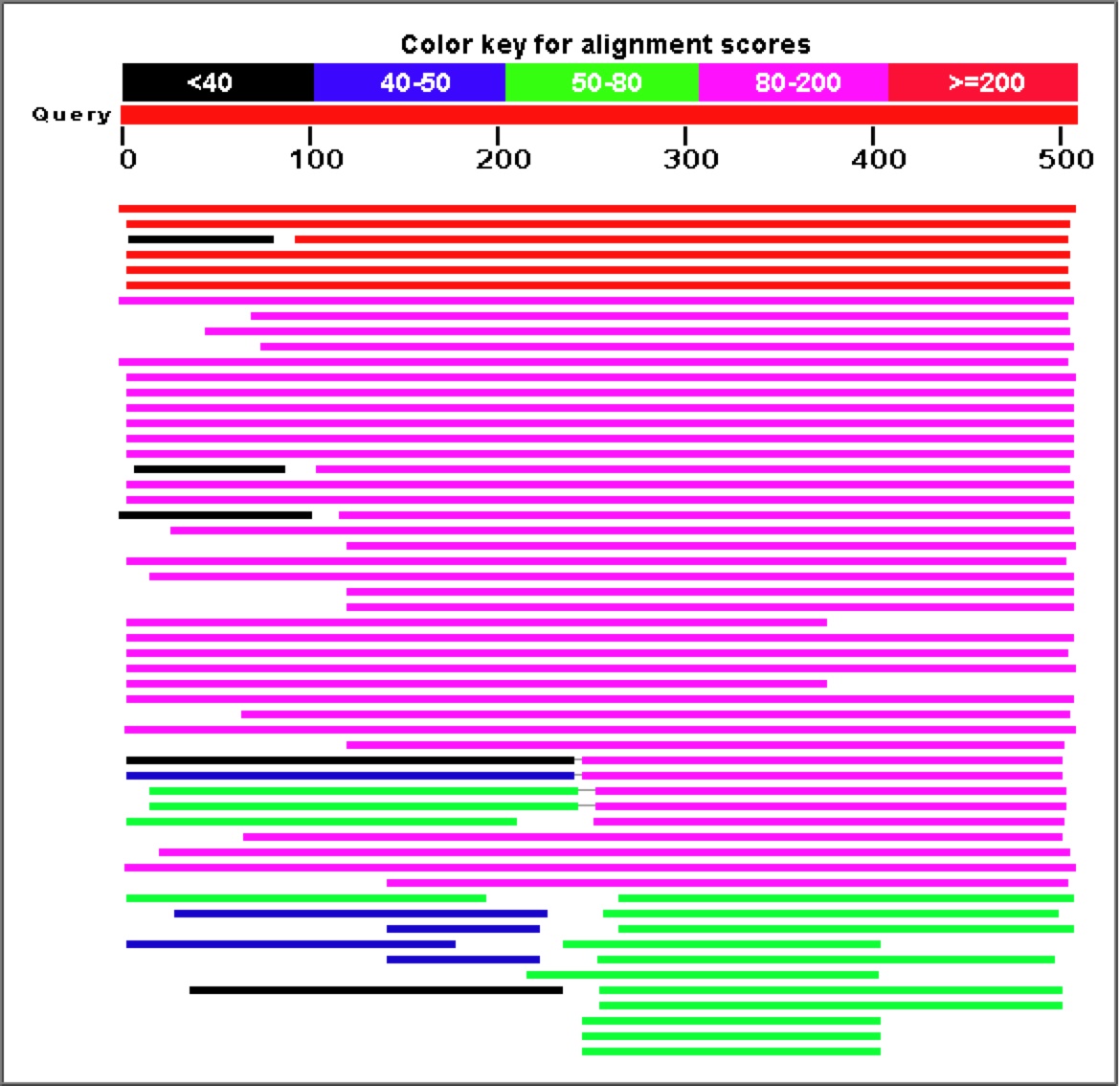

Figure 7. A Blastp search for proteins with similar sequences to YGR093w's predicted polypeptide sequence produces a large number of highly similar proteins.
Once again, proteins involved in mRNA splicing and potentially part of the spliceosome (CwfJ-family proteins) show a large amount of sequence similarity to YGR093w predicted protein. For this reason, my working hypothesis has been that YGR093w is also involved somehow in mRNA splicing, perhaps as a subunit of the spliceosome.
DIP - This site had a node assigned YGR093w, but identified no other proteins that interact with this protein.
ExPASy 2D Gel - No spots on the yeast 2D protein gel were identified as YGR093w.
ExPASy Database - Of all databases linked to the ExPASy search engine, only the UniPro Knowledgebase produced any information about YGR093w. However, this information only repeated data already covered on MFYG webpage, such as sequence and conserved domain data rehashed earlier on this page.
TRIPLES NORF, KEGG, Enzymes and Metabolic Pathways, PROWL, PDB, and Y2H - All contained no information on YGR093w's protein structure, function, or interactions with other proteins.
Still under development, current proteomic databases have many gaps of information. Even for a species as simple and well-characterized as yeast (Saccharomyces cerevisiae), many genes and potential genes (ORFs) have not been fully or even partially catagorized by the several available methods. To determine whether YGR093w is an actual gene and what function(s) its protein performs, I would use each common assay. This assays include: yeast 2-hybrid screening, LacZ transposon insertion, 2D gel/MS, and ICAT under several conditions.
First, I would like to determine whether or not the predicted YGR093w product can be isolated from yeast cells. Using software tools, like those provided on PROWL, a researcher can get a rough prediction of the final protein product's MW and isoelectric point. A 2D gel separates proteins by these characteristics, so it would make sense to run all yeast proteins on a 2D and look in the predicted area for YGR093w product. Analyzing the spots on the 2D gel with MS, I could easily determine whether the dot represented my predicted protein product, as well as the exact sequence of real protein product.
From this point, I would use LacZ transposon insertion and ICAT methods to gather more data about YGR093w's cellular component and importance to normal yeast activity. The TRIPLES database contains information on clones with transposon insertions in various yeast genes. Some genes have several clones with insertions at different places within there coding region, but many more have no clones whatsoever. This methodology can give a rough idea of where the gene product is normally localized. It has been reported that YGR093w product is found in the nucleus, but more supporting data would lend credence to these findings. By testing the effects of this disruption on general yeast growth, I could get an idea of how important YGR093w is to essential yeast activities. If it functions in mRNA splicing, I would expect to see YGR093w protein localized in the nucleus. I would also expect that the transposon insertion would have a very strong effect on overall yeast growth. ICAT methodology provides a way for researchers to detect varying levels of specific protein production under varying conditions. In order for this method to be applicable, it must be possible to grow the sample cells under various controlled conditions. This is possible with the ever-useful model organism, yeast. So, I would use ICAT under various experimental conditions (i. e. mutagens, ethanol, glucose, heat, etc.) to determine their effects on production of YGR093w protein. If any of these conditions had a drastic effect on protein production, the difference between experimental and control protein levels would be easy to detect on MS. Potentially, LacZ should react similarly under these experimental conditions to other, annotated spliceosome proteins.
Yeast two-hybrid screens are a useful technique for identifying protein-protein interactions. If YGR093w indeed comprises part of the spliceosome, it most likely interacts with many other spliceosome proteins. Yeast two-hybrid analysis with YGR093w as the bait protein would help determine if YGR093w actually interacts with any such proteins. This method is not perfect, as it may not identify all proteins that interact with YGR093w, but it still provides an important first step. If YGR093w does not interact with any known/suspected spliceosome proteins, this would force me to rethink my hypotheses about YGR093w's biological process and molecular function.
References
Chatton B, Walter P, Ebel J, Lacroute F, and Fasiolo F,. 1987. The Yeast Vas1 Gene Encodes Both Mitochondrial and Cytoplasmic Valyl-tRNA Synthetases. J Biol Chem. 261 (1): 52-57.
Kumar, A., Cheung, K.-H., Ross-Macdonald, P., Coelho, P.S.R., Miller, P., and Snyder, M. (2000). TRIPLES: a Database of Gene Function in S. cerevisiae. Nucleic Acids Res. 28, 81-84. <http://ygac.med.yale.edu/triples/basic_search.asp>. Accessed 2005 November 16.
Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWeese-Scott C, Geer LY, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Liebert CA, Liu C, Lu F, Marchler GH, Mullokandov M, Shoemaker BA, Simonyan V, Song JS, Thiessen PA, Yamashita RA, Yin JJ, Zhang D, Bryant SH. 2005. "CDD: a Conserved Domain Database for protein classification.", Nucleic Acids Res. 33: D192-6. <http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi>. Accessed 2005 November 17.
Xenarios I, Rice DW, Salwinski L, Baron MK, Marcotte EM, Eisenberg D (2000) DIP: The Database of Interacting Proteins. NAR 28 : 289-9. <http://dip.doe-mbi.ucla.edu/dip/Search.cgi?SM=3>. Accessed 2005 November 16.
Links
Davidson College Biology Department
© 2005 Department of Biology, Davidson College, Davidson, NC 28036
Please direct comments, criticisms and questions to andrysdale "at" davidson.edu