
Teaching Chips are designed to make DNA microarrays easier to bring into the teaching laboratory. Teaching Chips:

Sample teaching chip where 10 genes were probed simultaneously. No RNA or genomic DNA required. Each gene was spotted in duplicate and there are many spaces where no DNA was printed. Each gene was also spotted out at different concentrations to test the effect this would have. You can see the spots that smeared have the highest DNA concentration - more than could bind to the glass. Data produced by Emily Oldham ('03) of Davidson College.
Teaching chips are printed with double stranded DNA targets on glass slides using a new method that gives improved signal.
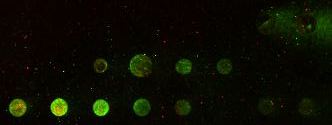
Second generation chip comparing two different ways of preparing the DNA for spotting. Only one gene's probe was used and the gene was spotted at different concentrations to produce different intensities of signal. Also, two different negative controls were included and these spots are all blank.
The new way produced brighter spots and cost much less to produce. We will be testing this method for all 11 genes (10 experimental yeast genes and 1 fly gene as negative control. Data produced by Dan Pierce ('03) of Davdison College.
Eleven genes (10 yeast genes and one negative control Drosophila gene) are printed at different concentrations.
The probes are oligonucleotides that are detected with Genisphere's 3DNA (dendrimers only; saves money).
Many experiments can be simulated based on published results.
Students can conduct research on the reliability of DNA microarray data collection and analysis.
Data analysis can be performed with MAGIC Tool software.
© Copyright 2003 Department of Biology, Davidson College, Davidson, NC
28035
Send comments, questions, and suggestions to: macampbell@davidson.edu