The gene SSK1 is found on chromosome 7 in Saccharomyces cerevisae. The protein it encodes is found in the cytoplasm and is part of a two-component osmosensor that regulates a MAP kinase cascade in response to hyperosmotic stress; when activated, SSK1 induces the MAP kinase cascade to raise the level of glycerol in the cell to restore osmotic balance. For more information about this gene, see My Favorite Yeast Genes and My Favorite Yeast Expression.
SSK1 is a 78.47 kDa protein with a pI of 6.8 (PROWL, 2004). The amino acid sequence of SSK1, translated from the gene's coding sequence, was obtained from the Saccharomyces Gene Database (SGD), as seen in Figure 1. A search of the Protein Data Bank (PDB) yielded no results for a 3-D structure of SSK1 (PDB, 2004), but the Pfam database contained a graphical representation of the domain structure of SSK1, as seen in Figure 2. The response regulator domain is the site of interaction with the other proteins in the two-component osmoregulatory response system (Pfam, 2004). There were no recorded data for SSK1 in the Yeast Resource Center's Two-Hybrid Analysis Database (YRC Two-Hybrid Analysis, 2004) or the Swiss 2-D Gel Page (Swiss 2-D Page, 2004).

Figure 1: The amino acid sequence of SSK1 (SGD, 2004).

Figure 2: Pfam entry for SSK1 (Pfam, 2004).
The TRIPLES database contains information about disrupted phenotypes caused by the insertion of transposons in various genes. As seen in Figure 3, scientists have produced several "clones" in which a transposon disrupts the gene SSK1. In the case of most of these clones, expression levels as measured by LacZ coloring intensity were equal in both vegetative and sporulation growth conditions. Expression levels were slightly higher during vegetative growth in the clones TN7-6D4 and TN7-13H3 and slightly higher during sporulation for the clone V53B12 (TRIPLES, 2004). For the most part, expression levels were fairly low, consistent with DNA microarray data showing that SSK1 is repressed during sporulation, but the clone TN7-24G4 was an "intense blue" in both conditions, as seen in Figure 4, so perhaps the transposon in that clone disrupted a site within the gene to which a repressor protein binds. There was further information on the disrupted phenotypes of clones V52H2, V54D10, and V53B12. As seen in Figure 5, the clone V52H2 exhibited a strong difference in growth in the 003SDS assay (YPD + 0.003% SDS); differences in growth were measured by a comparison of growth under these conditions to growth of the same clone on YPD. The clone V54D10 also exhibited a strong growth difference in the 003SDS assay (Figure 6), as did the clone V53B12 (Figure 7). The 003SDS assay results "indicate a role in cell wall maintenance and biogenesis," which is somewhat surprising given that SSK1 is a cytoplasmic protein involved in the osmotic stress response, although osmoregulation does affect the cell wall. As I have suggested before (see My Favorite Yeast Expression), SSK1 could be involved in cytoskeletal structure and organization and thus have an effect on the maintenance of the cell wall; however, no differences in growth were observed in the 10Ben assay (YPD + 10mg/mL benomyl), which would have indicated a role in microtubule dynamics, nor were any differences in growth observed under other "cell wall maintenance" conditions. SSK1 is not, therefore, crucial for cell wall maintenance. It is puzzling that the clones also exhibited no differences in growth in the 9NaCl assay (YPD + 0.9M NaCl), which was a test of salt-sensitivity. There may be an alternate response pathway for salt-stress that doesn't involve the SSK1 system, or SSK1 may be involved in responses only to specific stressors and not osmotic stress in general.

Figure 3: TRIPLES database results for SSK1 (TRIPLES, 2004).
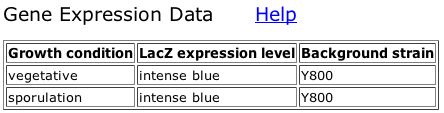
Figure 4: Expression data for clone TN7-24G4 (TRIPLES, 2004).

Figure 5: Disruption phenotype data for clone V52H2 (TRIPLES, 2004).

Figure 6: Disruption phenotype data for clone V54D10 (TRIPLES, 2004).

Figure 7: Disruption phenotype data for clone V53B12 (TRIPLES, 2004).
The Database of Interacting Proteins (DIP) produced a rather elaborate graphical map of a network of interactions between SSK1 and other proteins, as seen in Figure 8. SSK1 is known to interact with several proteins, including YPD1 and SSK2, both of which are denoted by orange nodes connected to SSK1 by green lines indicating verified interactions in the lower right of the graph. Unverified interactions with the proteins EST1 (telomerase length regulation), QRI8 (protein degradation), SAS10 ("something about silencing"), and HEX3 (DNA damage response) are indicated by red lines. The possible SSK1-HEX3 interaction, in particular, is of note due to its correlation with DNA microarray data suggesting that SSK1 plays a role in the DNA damage response as well as the osmotic stress response (HEX3 and SSK1 had similar expression patterns under certain conditions; see My Favorite Yeast Expression for further information). SSK1 also interacts with the protein CDC28, denoted by the orange node surrounded by many yellow nodes in the upper left of the graph. The catalytic subunit of the main cell cycle cyclin-dependent kinase (CDK), CDC28 helps regulate transitions between cell cycle and mitotic phases (SGD, 2004). Perhaps it regulates SSK1 activity during mitosis (i.e. SSK1 is a potential CDC28 substrate and is inactivated through phosphorylation by CDC28); DNA microarray data shows that SSK1 is strongly repressed during sporulation, possibly to conserve energy during reproduction, so perhaps it is also repressed during certain points in the cell cycle, such as asexual reproduction, for the same reasons--to conserve energy until the cell resumes normal activity.

Figure 8: Graphical map of SSK1's interactions with other proteins. The red node is SSK1. Green lines denote verified interactions; red lines denote unverified results of high-throughput interaction screens. Line width indicates the number of experiments identifying the interaction (DIP, 2004).
Further experiments could reveal much about other roles of SSK1 within the cell, some of which have already been suggested by DNA microarray data and protein databases. The yeast two-hybrid method has not yet been used to analyze SSK1; I would be interested in further verification of the interaction between SSK1 and HEX3 using SSK1 as the bait protein as part of an investigation into a possible role of SSK1 in the DNA damage response. I would also suggest determining more about SSK1's activity during various stages of the cell cycle; investigation of the connection between SSK1 and CDC28 could be a good starting point. As I have not found a comprehensive list of stressors to which SSK1 responds, but only possibilities based on DNA microarray data, I would like to see phenotype macroarrays testing the survival ability of SSK1 mutants or knockouts upon exposure to a wide variety of osmotic stresses. Further experiments with the kinase functionality high-throughput method could also be useful by revealing to which substrates SSK1 will bind, which could help verify SSK1's involvement in more than one response pathway or biological process. Finally, determining the protein's structure would greatly help any investigation into how it achieves its purpose within the cell.