MacDNAsis
Holding It Together:a Closer
Look at Collagen
Using the MacDNAsis program (version
3.5), several different analyses of the cDNA for collagen were performed:
- the largest open reading frame
(ORF) was obtained from human collagen cDNA,
- the nucleotide sequence from the
largest ORF was translated and a predicted molecular weight for human collagen
was determined,
- a Kyte and Doolittle analysis was
performed on the translated ORF to create a hydropathy plot in hopes of
determining if collagen is an integral membrane protein,
- a Hopp and Wood analysis was performed
on the translated ORF to create an antigenicity plot to determine the portions
of the protein against which a monoclonal antibody could be developed,
- based on the ORF from human collagen,
the predicted secondary structure of collagen was determined and compared
to the three dimensional Rasmol image obtained from the NCBI archives,
- a multiple sequence alignment was
performed on the primary protein structure of collagen from five different
organisms: Gallus gallus (chicken), Caenorhabditis elegans
(worm), Drosophila melanogaster (fly), Mus musculus (mouse),
and Homo sapiens (human),
- a phylogenetic tree was constructed
to determine the degree of amino acid conservation between these five species
over time.
Part I: Open Reading Frame
(ORF)
The DNA analyzed was the human cDNA
for collagen. The largest reading frame of this DNA was chosen. This segment
of the DNA began at nucleotide number 235 and terminated at nucleotide
5271. The image below shows the entire segment of cDNA. The largest reading
frame, designated by the black box was the segment of DNA used for the
rest of the DNA analysis.
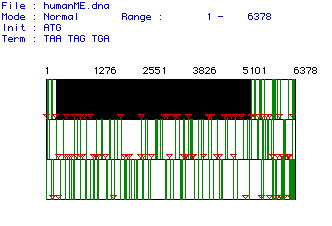
Figure #1: Determination of the Largest Open Reading Frame
(ORF). This image shows the three possible reading frames for the collagen
cDNA isolated from humans. The nucleotide numbers are listed above the
reading frames. Red triangles indicate start (AUG) codons while vertical
green lines represent stop codons. The largest ORF was found in the first
open reading frame, starting at nucleotide 235 and terminating at nucleotide
5271.
The entire collagen cDNA sequence (along with protein
translation) was obtained from the NCBI (National Center for Biotechnology
Information. 14 November 1997. Entrez Protein Search. <http://www.ncbi.nlm.nih.gov>
Accessed: 20 March 1998) and can be viewed by clicking below:

Homo
sapiens
Part II: Translation and Determination
of the Predicted Molecular Weight
The selected ORF (from above) was
translated (DNA to protein) and its predicted molecular weight was then
calculated (in daltons):
162,452.55
Part III: Hydropathy Plot
(Kyte and Doolittle)
Using the algorithm developed by
Kyte and Doolittle, a hydropathy plot was made to determine if collagen
is an integral membrane protein. Below is the plot:

Figure #2: Hydropathy Plot. This plot was constructed
using the algorithm developed by Kyte and Doolittle. A reading of greater
than or equal to +1.8 indicates an area of the protein that is hydrophobic
enough to be a transmembrane domain. No peak on this plot has a hydrophobicity
greater than or equal to +1.8, indicating that there are no transmembrane
domains.
Positive values on this plot indicate areas of the protein
that are hydrophobic. To be a candidate for a transmembrane domain, a segment
of the protein must have a hydrophobicity reading greater than or equal
to +1.8. As indicated in this plot, there are no portions of collagen with
a hydrophobicity greater than or equal to +1.8. There are, however, several
peaks that come close to the +1.8 hydrophobic threshold, including a peak
at the very beginning of the plot.
Collagen is primarily an extracellular protein. To be
secreted, it must be translated into the ER and then modified in the Golgi
apparatus. In order to find its way into the ER, collagen carries a hydrophobic
signal sequence at its 5' end. This sequence binds the ribosome and the
collagen mRNA (that is being translated) to the ER membrane so that the
mRNA can be translated directly into the lumen of the ER. The hydrophobic
peak at the left-most part of Figure #2, therefore, may correspond
to a signal sequence.
With the exception of this peak, there are no other peaks
with hydrophobicity readings great enough to be transmembrane domains suggesting
that collagen is not an integral membrane protein.
Part IV: Antigenicity Plot
(Hopp and Wood)
Using the algorithm developed by
Hopp and Wood, an antigenicity plot was constructed. The following are
the results of the antigenicty study:
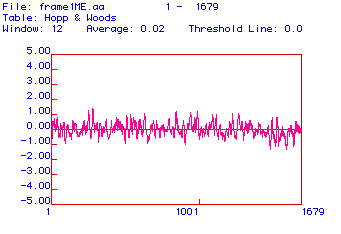
Figure #3: Antigenicity Plot. This plot was constructed
using the algorithm developed by Hopp and Wood. Positive values indicate
hydrophilic or antigenic areas.
Antigenic plots are used to determine areas of the protein
that are charged and therefore hydrophilic. Charged areas of the protein
cannot be associated with the phospholipid bilayer (because it is hydrophobic);
therefore, these areas of the protein point away from the membrane. In
this configuration, these segments of the protein can be recognized and
bound by immunoglobulins (antibodies). To make a monoclonal antibody against
collagen (to be able to detect it), one of these areas would be used. The
antigenicity plot above reveals numerous antigenic (hydrophilic) areas
that could be used for monoclonal antibody production.
Part V: Predicted Secondary
Structure
The translated ORF of the human
collagen cDNA was further studied by constructing a predicted secondary
structure map:
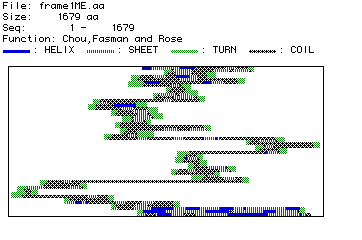
Figure #4: Predicted Secondary Structure of Collagen.
Based on the position of amino acid residues, their side chains, and
their associated hydrogen and oxygen molecules (which hydrogen bond to
form secondary structure), a plot of predicted secondary structure was
constructed.
This predicted secondary structure can be compared to
the actual, crystallized structure of type VI collagen. Click below to
see the Rasmol image of type VI collagen. Once the image has appeared,
click on Display and then drag down to Ribbons. This is the
best format to see the secondary structure (alpha helices and beta-pleated
sheets) of collagen to which the above map can be compared.
The predicted secondary structure from the MacDNAsis agrees,
in part, with the actual three dimensional structure depicted in the Rasmol
image. The Rasmol image is not, however, the complete type VI collagen
protein. Instead, it is only a fragment. Through a comparison with the
predicted secondary structure determined by the MacDNAsis program, it appears
that the Rasmol image shows the first third of the total collagen protein.
Both the predicted secondary structure and the Rasmol image both start
with an alpha helix followed by beta sheets and interspresed coiled sections
of protein. The Rasmol image terminates with a coiled section followed
immediately by a long alpha helix. This same pattern appears in the predicted
secondary structure nearly a third of the way through the map.
Furthermore, by clicking on Windows and then Command
Line (of the Rasmac program), some facts are given about the Rasmol
image of type VI collagen. Some of the relevant information with respect
to this discussion is:
- Number of alpha helices: 2
- Number of strands: 5
- Number of turns: 7
These numbers give further credence to the idea that the
Rasmol image graphically represents the first third of the predicted secondary
structure. The first seven turns of the Rasmol image correspond (in terms
of the postion and number of alpha helices, strands, beta pleated sheets,
coils, and turns) almost exactly with the first third of the structure
predicted by the MacDNAsis.
Finally, there are many types of collagen (thus far at
least twelve different types of collagen have been reported in the human
body). Therefore, some minor discrepancies between the Rasmol image (type
VI collagen) and the MacDNAsis predicted secondary structure (type IV collagen)
may be present because the human collagen DNA used in the MacDNAsis is
not be the same type of collagen that appears in the Rasmol image.
Part VI: Multiple Sequence
Alignment
This portion of the analysis consisted of comparing the
collagen protein sequences (primary structure or order of amino acid residues)
from each of the five organisms listed above (Gallus
gallus, Caenorhabditis elegans,
Drosophila melanogaster, Mus musculus, and Homo sapiens).
A short sequence of the comparison appears below. It is clear that the
collagen samples contain a limited amount of homology as the amino acid
sequences had to be manipulated (spaces were added or removed) quite extensively
in order to obtain the best alignment. Normally, this would suggest poor
homology between the protein samples (i.e., the proteins most likely
do not have a common protein origin or "ancestor"). In this case,
however, the protein from Gallus gallus (chicken) was only a small
piece or fragment of the entire protein. Because the entire protein was
not available for analysis, the entire alignment procedure was altered.
The poor alignment, therefore, might be explained in this manner. Furthemore,
the collagen protein sequences from worm and fly were also quite short
making an accurate (or exact) alignment very difficult.
The cDNA (and translated protein)
sequences for each organism's collagen can be viewed by clicking next to
the appropriate image below:
 Gallus
gallus
Gallus
gallus Drosophila
melanogaster
Drosophila
melanogaster Mus
musculus
Mus
musculus Caenorhabditis
elegans
Caenorhabditis
elegans Homo
sapiens
Homo
sapiens
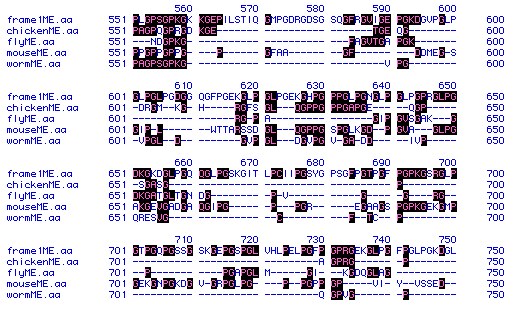
Figure #5: Multiple Sequence Alignment. A multiple
sequence alignment was performed on the amino acid sequences of collagen
from the five organisms listed above (Gallus gallus,
Caenorhabditis elegans, Drosophila melanogaster, Mus musculus,
and Homo sapiens). Part of the alignment is shown here. Residues
with black boxes indicate those residues that appear in more than one organism.
The frame1ME.aa segment is the collagen protein sequence
from the human. Spaces (-) were introduced in order to improve alignment
between the five different collagen sequences.
Part VII: Phylogenetic Tree
In order to obtain a quantitative
description of the alignment (above), a comparative, phylogenetic analysis
was performed between the five types of collagen. The goal of this analysis
was to determine the degree of amino acid conservation. The percentages
listed in the tree tell the likelihood that the observed overlap between
two sequences was due to a common protein origin versus chance.
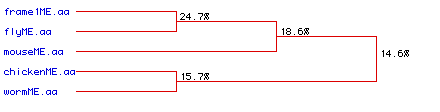
Figure #6: Phylogenetic Tree. A phylogenetic analysis
was performed to determine the degree of amino acid conservation between
the five types of collagen over time. Percentages indicate the likelihood
that the overlap observed between different sources of collagen are from
the same ancestral origin. Frame1ME.aa refers to the collagen
protein from human.
This image supports the conclusion obtained above through
the alignment study (Part VI: Multiple Sequence
Alignment): very little homology exists
between the different collagens (from different organsisms). This conclusion
is not definative; because a fragment from the chicken was used and because
the fly and the worm collagen samples were so short, the alignment/homology
analysis is probably not perfect.
The cDNA (and translated protein)
sequences for each organism's collagen can be viewed by clicking next to
the appropriate image below:
 Gallus
gallus
Gallus
gallus Drosophila
melanogaster
Drosophila
melanogaster Mus
musculus
Mus
musculus Caenorhabditis
elegans
Caenorhabditis
elegans Homo
sapiens
Homo
sapiens
Return
to Personal Homepage
Return to Davidson College Molecular Biology
Home Page
Send comments, questions, and suggestions to:
mjayellis@aol.com
