MacDNAsis analysis of Cytochrome C Oxidase
Subunit II
After collecting the cDNA and amino acid sequences for the five genome
organisms from Genbank (see this page for details),
I used the cat cDNA sequence for analysis with the MacDNAsis sequence analysis
software. The results for the analysis are presented below.
Open Reading Frame (ORF) search
MacDNAsis was used to search the cat cDNA sequence for open reading frames.
The search showed that the largest open reading frame was in the frame
starting with the first base pair in the cDNa sequence, and was from base
pair1 to base pair 137. The search results are shown in Fig. 1 below.
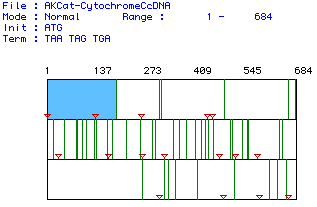
Figure 1. Search results for cat Cytochrome C Oxidase Subunit II open
reading frames using the MacDNAsis program.Standard start and stop codons
were used for the search. The red inverted triangles represent start codons
and the vertical green lines represent the stop codons. The blue area marks
the largest open reading frame found, from base pair 1 to base pair 137.
Protein sequence and molecular weight
The MacDNAsis program was used to translate the largest open reading frame
in the cDNA (see above) into the corresponding amino acid sequence, and
calculate the number of occurrences of each amino acid and the molecular
weight of the the protein. The results of the translation are shown in
Figure 2 below.
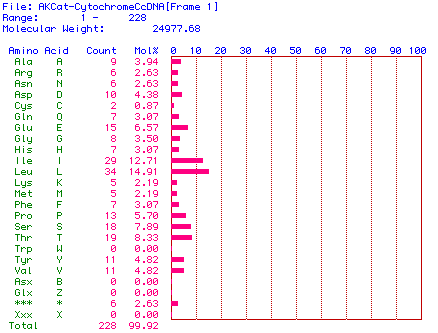
Figure 2. Amino acid content of the protein sequence translated from
the cat Cytochrome C Oxidase Subunit II cDNA sequence. This shows
the molecular weight as well as the concentration of amino acids.
The molecular weight of the protein sequence was calculated to be 24977.68
daltons, or about 24.98 kDa.
Hydropathy plot of amino acid sequence
A hydropathy plot for the translated protein sequence was done using the
Kyte and Doolittle algorithm. The results are shown in Fig.3.
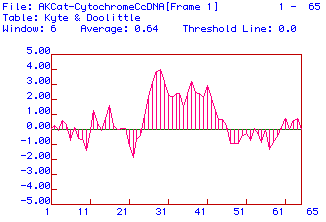
Figure 3. Hydropathy plot for protein sequence translated from cat
Cytochrome C Oxidase Subunit II cDNA using the Kyte and Doolittle algorithm.
A window of 6 amino acids was used and the threshold for a transmembrane
domain was 0.0.
The hydropathy plot shows that there is possibly 10 hydrophobic areas
of this subunit protein. These could be areas that are inside the
protein when it is in its tertiary structure. Since Cytochrome C
Oxidase Subunit II is a subunit, these hydrophobic areas may also be hidden
beneath the other subunits.
Antigenicity analysis
An antigenicity plot was generated for the translated protein sequence
using the Hopp and Woods algorithm. The plot is shown in Fig. 4.
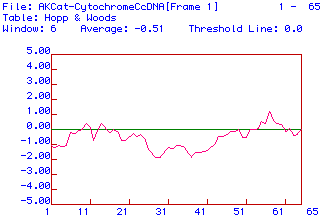
Figure 4. Antigenicity plot for protein sequence translated from Cat
Cytochrome C Oxidase Subunit II cDNA. The algorithm used was that
of Hopp and Woods and the window used was 6 amino acids.
The Hopp & Woods method of predicting secondary structure paints
a similar picture of the protein. This shows the hydrophilic and
highly charged areas of the polypeptide. There does not appear to
be more than three hydrophilic and highly charged areas.
I think that a likely place to use for monoclonal
antibody generation would be the areas between 55 and 61 because they are
the most hydrophilic. If one wanted to just generate monoclonal antibodies
for Cytochrome C Oxidase, I would suggest using a more hydrophilic subunit
(I or III).
Secondary structure prediction
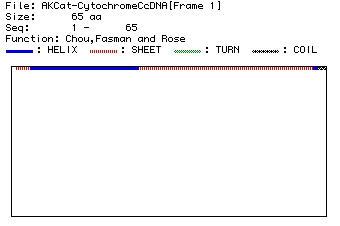
I used the Chou, Fasman and Rose algorithm to predict the secondary structure
of the protein using the primary sequence. The results are shown in Fig.5.
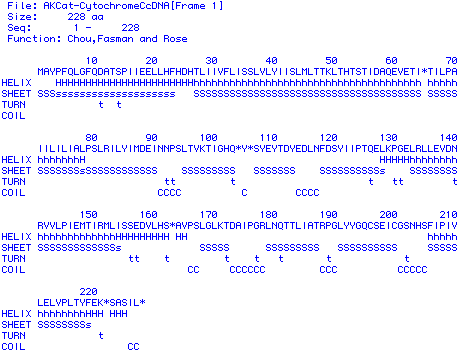
Figure 5. Secondary structure prediction for protein sequence translated
from Cat Cytochrome C Oxidase Subunit II cDNA. The algorithm used was that
of Chou, Fasman and Rose. The "H" strings mark helical structure, the "S"
strings mark sheets, the "t"s mark turns and the "C"s mark coils.
Fig. 6. Secondary Structure Prediction
Figure 6. This shows another view of the Chou, Fasman and Rose secondary
structure prediction.
The secondary structure predicted is fitting for a subunit protein.
The easiest way to picture the protein is by using the Rasmol
image of Cytochrome C Oxidase Subunit II. I would
also recommend looking at this protein as it interacts with the other cytochrome
c oxidase subunits. Rasmole image of entire Cytochrome
C Oxidase. This will also help to reveal the possible reasons
for the many hydrophobic regions of this subunit (such as subunit binding
sites).
Multiple sequence alignment and phylogenetic tree
The Genbank amino acid sequences for Cytochrome C Oxidase Subunit II from
the five genome organisms was used to perform a multiple sequence alignment
test for sequence homology. The original protein sequences can be found
here:
Yeast (Saccharomyces cerevisiae)
Horse (Equus caballus)
Frog (Xenopus laevis)
Tarsius (Tarsius syrichta)
Cat (Felis catus)
The results of the alignment are shown in Fig.7.

Figure 6. Multiple alignment results for the amino acid sequences for
Cytochrome C Oxidase Subunit II from the five genome organisms. The consensus
sequences are highlighted in black.
The multiple sequence alignment results show that a large proportion
of the amino acids in the primary sequence of the three mamalian species
and amphibian match up very closely. This indicates that cytochrome c oxidase
subunit (and most likely the whole protein) has been conserved through
evoultion.
I also used the MacDNAsis program to generate a phylogenetic tree based
on the sequence homology of Cytochrome C Oxidase Subunit II. The tree is
shown in Fig. 8.
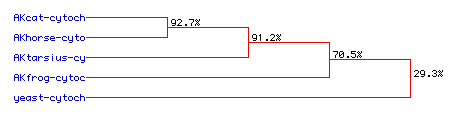
Figure 7. Phylogenetic tree based on the sequence homology of Cytochrome
C Oxidase Subunit II for Cat, Horse, Tarsius, Frog, and Yeast. The
estimated percentage homology with the Cat amino acid sequence is indicated
on each branching point.
[Home] [Back
to the Davidson College Molecular Biology page]
Any comments or questions can be answered by e-mailing me at ankazama@davidson.edu