* This
webpage was produced as an assignment for an undergraduate course at Davidson
College*
MacDNasis Results for Enolase
Enolase cDNA from 5 different
species was analyzed using MacDNAsis software. The results from the analysis
of cDNA from Homo sapiens is shown in the following figures.
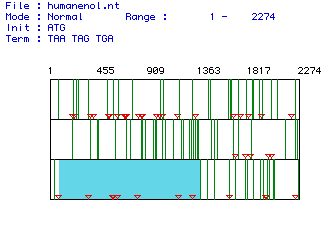
I.
Determination of the largest open reading frame (ORF):

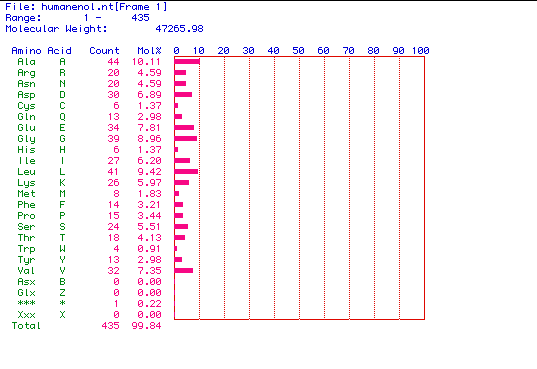
II.
Molecular weight prediction for enolase:
III.
Kyte and Doolittle hydropathy plot of enolase:
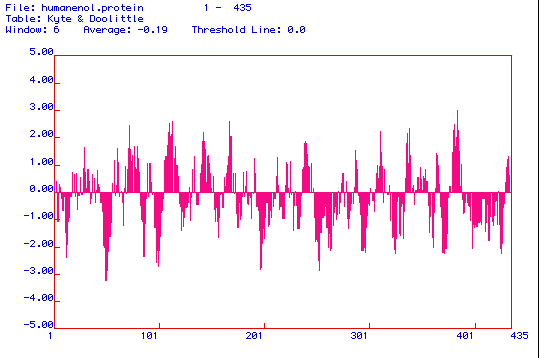
Figure 3. The above image is a Kyte and Doolittle hydropathy plot of enolase
that was produced using MacDNAsis software. Kyte and Doolittle hydropathy
plots are used to determine whether or not a protein is an integral membrane
protein. The amino acid regions that are greater than 0 on the plot are
hydrophobic regions, and the areas less than 0 indicate hydrophilic amino
acid regions. If a peak reaches a value of positive 2, than the protein
is considered to be an integral membrane protein. When analyzing the hydropathy
plot above, it appears that enolase is an integral membrane protein with
approximately 7 transmembrane regions.
IV.
Hopp and Woods antigenicity plot for enolase:
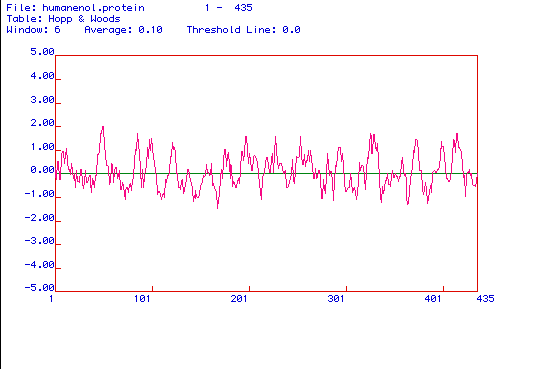
Figure 4. The above image is a Hopp and Woods
hydrophilicity plot of enolase. A Hopp and Woods plot is used to predict
hydrophilic regions in a protein. The amino acid regions that appear to
have the most positive values on the plot are the most hydrophilic regions
of the protein. Knowing which amino acids are the most hydrophilic, one
can determine which portion of the protein should be used to create a peptide
and a monoclonal antibody against that peptide. When analyzing the antigenecity
plot above, it appears that the best portions of enolase from which a peptide
and a monoclonal antibody should be generated are around amino acids 30-50,
towards the middle of the protein at about amino acids 250-320, or at the
end of the protein that includes the amino acids 400-430 because these
are the most hydrophilic portions of the protein.
V. Secondary structure of enolase:
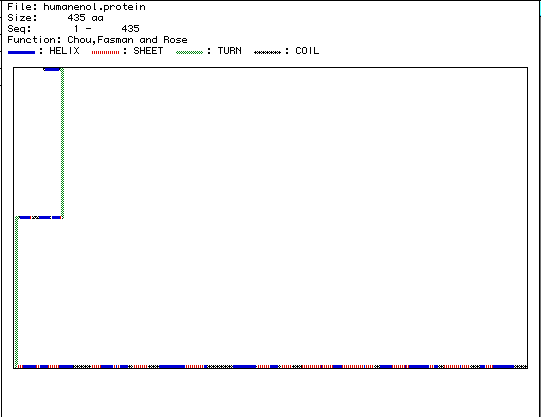
Figure 5. The above image is the secondary
structure of enolase that was generated using MacDNasis. According to this
figure, enolase is composed of 24 alpha helices, 22 beta-pleated sheets,
2 turns, and 15 coils. If you would like to view the 3-D Rasmol structure
for enolase click
here.
VI. Multiple amino acid sequence alignment of enolase from 5 different species:
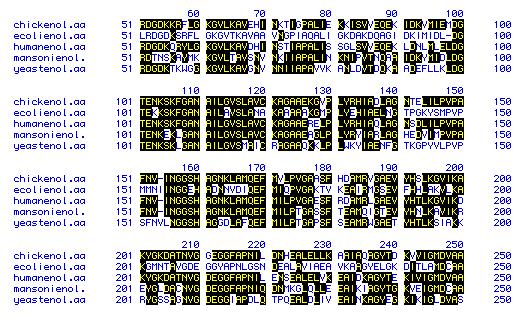
Figure 6. This figure shows the amino acid
sequence similarity between enolase from 5 species: Escherichia coli,
Schistosoma mansoni, Homo sapiens, Gallus gallus,
and Schizosaccharomyces pombe. The amino acids that are shaded in
black indicate those sequences that are similar, and those amino acids
that are not shaded indicate amino acids that deviate from the other species'
amino acids at those portions of the protein. Because the majority of the
figure is shaded, a high degree of sequence similarity exits for enolase
among the 5 different species. To see the amino acid sequences generated
for the 5 species using Genbank, click on one of the following species:
Escherichia
coli, Schistosoma
mansoni, Homo
sapiens, Gallus
gallus, Schizosaccharomyces
pombe.
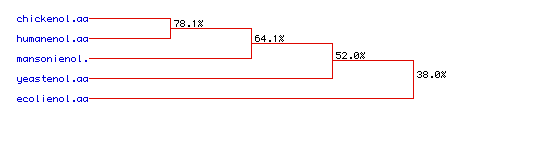
VII. Phylogenetic tree for enolase:
Click
here to return to Carrie's homepage
Click
here to go to the Davidson College Molecular Biology web page
Any
questions? E-mail me at: casmith@davidson.edu