
Figure 1: Figure legend from the DIP database
This web page was produced as an assignment for an undergraduate course at Davidson College
My Favorite Yeast Protein: RGT2 and YDL133W
The following legend can be used to examine figures from the DIP database:

Figure 1: Figure legend from the DIP database
RGT2:
Background information:
For more information about RGT2 please visit my previous web sites:
My Favorite Yeast Gene
My Favorite Yeast Expression

Figure 2: Image of RGT2 and its role in the cell. It can be seen in the figure that RGT2 is a membrane protein that is involved in signal transduction. This image was obtained from the MIPS database.
Protein Information:

Figure 3: This is an image of the protein interactions of RGT2 (red dot) with other proteins. The green line between RGT2 and Lys14p indicates a core interaction between these two proteins. This image was obtained from the DIP databse.
| Since there was a core interaction between RGT2 and Lys14p it can be assumed that these two proteins probably are involved in similar pathways or have similar biological or molecluar processes. To investigate the possible similarities between these two proteins I searched for more information about Lys14p. The results of my search can be found below. |

Figure 4: Information regarding Lys14 its molecular function, biological process and location within the cell. This information was found in the SGD databse.
| It is interesting to note that Lys14 and RGT2 do not have similar molecular functions, biological processes or locations within the cell. However, it is important to note that Lys14 is located on the same chromosome of RGT2 (chromosome IV). Even though these two proteins are seemingly unrelated they may have other characteristcs in common that are not evident at this time. |

Figure 5: This is an image of Benno figure 1. RGT2 is highlighted in this figure and some protein interactions can be seen. However, the interactions are not clear and it is not very useful in determining specific interactions between proteins. It is a good figure to see the proteome overall.
Future Experiments:
Most of the common experiments used in determining protein information have been performed on RGT2. However, a search of the Yeast 2 Hybrid (Y2H) database yielded no results. The Y2H experiment would be a good one to test the interactions between RGT2 and other proteins. By using this method of a bait and prey protein, more information could be obtained about specific interactions between RGT2 and other proteins. The ambiguity of the relationship between RGT2 and Lys17, for expample, may be resolved using this method. If this method shows an interaction between RGT2 and Lys17, there would be more evidence to support the conclusions made by the DIP database. |
YDL133W:
Background Information:
For more information about YDL133W please visit my previous websites:
My Favorite Yeast Gene
My Favorite Yeast Expression
Protein Information:

Figure 6: DIP graph representing protein interactions between YDL133W and several other proteins. It can be noted from this figure that there are several red interactions in this figure. A red line inducates that the interaction is unverified and the result of high-throughput methods of determining protein interactions. This type of interaction is less reliable than a green (core) interaction.
| Using the information included in the figure above, I searched for the GO functions of Far3p, Rmd1p, and Sec17p. The results of this search can be found below. |

Figure 7: DIP search results for Far3p.

Figure 8: DIP search results for Rmd1p.

Figure 9: DIP search results for Sec17p.
Using the information provided by the GO functions of the above proteins, I was not able to find out much more information about YDL133W. Like YDL133W, Rmd1p is a probable membrane protein but information regarding its molecular function, biological process and location in the cell were unknown. However, information about the other two proteins were inconclusive. Far3p is involved in cell cycle arrest in response to pheromone and is found in the ER. Sec17p is involved in ER to Golgi transport, is found peripheral to the membrane and its molecular process is that it is involved in NSP attachment. Since both Sec17p and Far3p are either involved in ER processes or are found in the ER, YDL133W may also have a role in Er processes. However, more experimentation is needed in order to determine if this is true. One other interesting thing to note is that in my previous web page on protein expression, I hypothesized that YDL133W may be involved in the sporulation process. Since Far3p is involved in cell cycle arrest, this hypothesis may not be too far off from the truth. The conclusion that YDL133W would be supportive of the interaction between Far3p and YDL133W. If YDL133W was in fact involved in sporulation, the cell would have to undergo cell cycle arrest. These two proteins may be more related than the DIP database indicates. |

Figure 10: This image shows the Benno results of protein interactions. It is hard to determine specific interactions between YDL133W and other proteins from this image. However, it is important to note that in this figure, YDL133W is directly attached by a gray line to YDL001W which is called Rmd1p in the DIP database. Since both sets of data yeilded the same interaction, this lends further support to the fact that YDL133W and YDL001W are in fact related or interact with eachother.
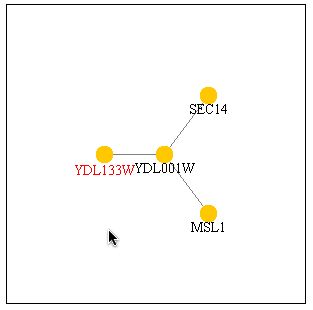
Figure 11: A search of PathCalling yielded the above graph. It appears that YDL133W and YDL001W have a strong interaction. Three databases have information that indicate a relationship between these two proteins. YDL001W is also known as Rmd1 which is required for sporulation.
| Since three databases have shown that YDL133W and YDL001W interact with eachother, it can be assumed that this interaction is indeed true. Since YDL001W in involved in sporulation and the databases indicate a strong interaction, it can be assumed that YDL133W is also involved in sporulation. My hypothesis from my previous web page has been confirmed by other databases indcluding DIP and the Benno figure. |

Figure 12: Y2H information regarding YDL133W. It is interesting to note that Sec17 is the bait protein used in this experiment. Sec17 was indicated to interact with YDL133W in the DIP database (Figure 6).
| The Y2H information above further suggests that YDL133W is indeed a membrane protein. Since SEC17 is involved in ER to Golgi transport and was the bait protein, YDL133W must travel through the ER and Golgi before finding its way to the plasma membrane. |
Future Experiments:
Since the celluar localization of YDL133W has been determined to be the plasma membrane, experiments need to be carried out which will help determine the molecular function and biological process of this protein. The biological process and molecular function of this protein could be determined by doing protein macroarray analysis of YDL133W. Also, knockout experiments could be carried out in which YDL133W specifically was removed from the genome and tests to see if the cell could induce sporulation would lend more insight into whether this protein does in fact regulate sporulation. These knockout experiments would also allow researchers to determine what aspects of the cell cycle, if any, YDL133W regulates. In addition, experiments in which both YDL133W and YDL001W were knocked out at the same time would help researchers determine what interactions they have with one another. By comparing the data of just the YDL133W knockout to the combined knockout of both YDL133W and YDL001W, researchers would be able to determine what differences occured and what interactions, if any, these two proteins have with one another. |
References:
Additional Y2H Results. 2004. <http://depts.washington.edu/~yeastrc/th_12.htm> Accessed 18 November 2004.
Benno Figure 1. 2004. <NB_Figure1color.pdf> Accessed 18 November 2004.
DIP Database. 2004.0< http://dip.doe-mbi.ucla.edu/dip/Search.cgi?SM=3> Accessed 18 November 2004.
MIPS Database. 2004. <http://mips.gsf.de/genre/proj/yeast/index.jsp> Accessed 18 November 2004.
PathCalling Database. 2004. <http://portal.curagen.com/extpc/com.curagen.portal.servlet.PortalYeastGene?geneIdIn=846&keywordIn=YDL133W> Accessed 18 November 2004.
Saccharomyces Genome Database. 2004. <http://www.yeastgenome.org/> Accessed 18 November 2004.
Back to Hompage
Davidson Department of Biology Homepage
Davidson Genomics
Davidson College
Questions or Comments?: e-mail me
© Copyright 2004 Department of Biology, Davidson College, Davidson, NC 28035