MacDNAsis Analysis
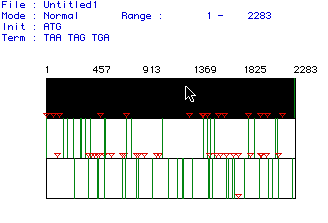 Figure 1. MacDNAsis analysis to determine the largest orf (open
reading frame) of Mus musculus DNA helicase protein. In the
figure, red triangles indicate start codons and green lines indicate stop
codons; the large black area with the arrow pointing to it is the largest
orf and consists of 2283 bp. To see the original Mus musculus
DNA helicase cDNA, click here.
Figure 1. MacDNAsis analysis to determine the largest orf (open
reading frame) of Mus musculus DNA helicase protein. In the
figure, red triangles indicate start codons and green lines indicate stop
codons; the large black area with the arrow pointing to it is the largest
orf and consists of 2283 bp. To see the original Mus musculus
DNA helicase cDNA, click here.
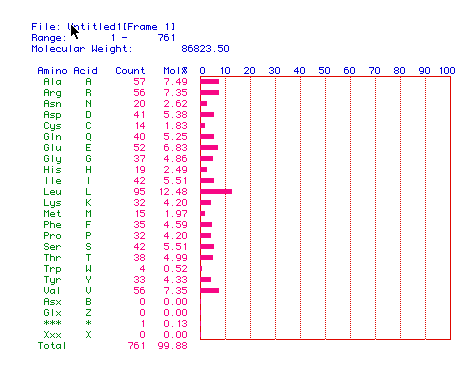 Figure 2. The translation of DNA helicase cDNA to protein.
From the figure one can determine the percent compostion of DNA helicase
by amino acid, and its molecular weight in daltons, which is 86823.50 daltons.
Figure 2. The translation of DNA helicase cDNA to protein.
From the figure one can determine the percent compostion of DNA helicase
by amino acid, and its molecular weight in daltons, which is 86823.50 daltons.
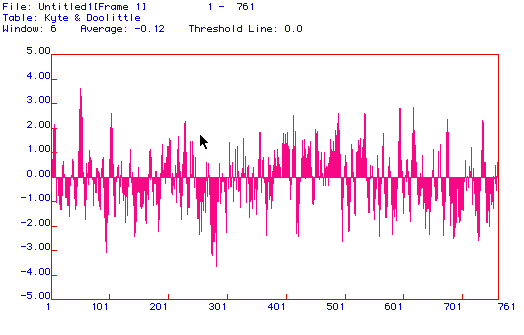 Figure 3. A hydropathy plot of DNA helicase using the Kyte and Doolittle
method. Though DNA helicase is not an integral membrane protein,
the hydropathy plot suggests that it is, as it contains many regions that
exceed the 2.00 limit. This discrepancy is most likely due to the
small size of the graphing window. If the window were increased one
would most likely find that the plot contains no regions that exceed the
2.00 limit, thus properly classifying DNA helicase as a non-integral membrane
protein.
Figure 3. A hydropathy plot of DNA helicase using the Kyte and Doolittle
method. Though DNA helicase is not an integral membrane protein,
the hydropathy plot suggests that it is, as it contains many regions that
exceed the 2.00 limit. This discrepancy is most likely due to the
small size of the graphing window. If the window were increased one
would most likely find that the plot contains no regions that exceed the
2.00 limit, thus properly classifying DNA helicase as a non-integral membrane
protein.
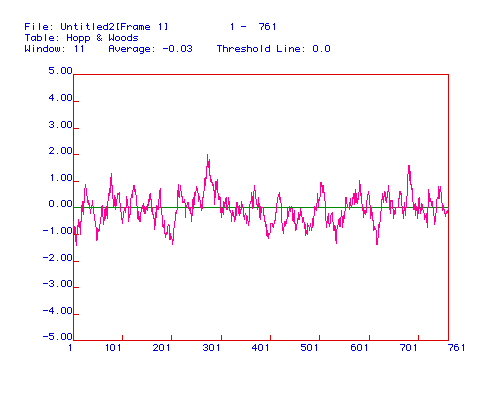 Figure 4. Antigenicity plot of Mus musculus
DNA helicase. The best region to use as a receptor for an antibody
would be the most hydrophilic regions of the protein, corresponding to
amino acid residues 275-285 and 685-692.
Figure 4. Antigenicity plot of Mus musculus
DNA helicase. The best region to use as a receptor for an antibody
would be the most hydrophilic regions of the protein, corresponding to
amino acid residues 275-285 and 685-692.
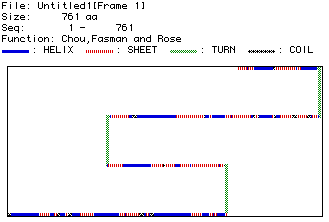 Figure 6. Representation of the secondary structure of mouse DNA helicase.
To compare this to the rasmol quarternary structure of DNA helicase in
Bacillus stearothermophilus, click here.
Figure 6. Representation of the secondary structure of mouse DNA helicase.
To compare this to the rasmol quarternary structure of DNA helicase in
Bacillus stearothermophilus, click here.
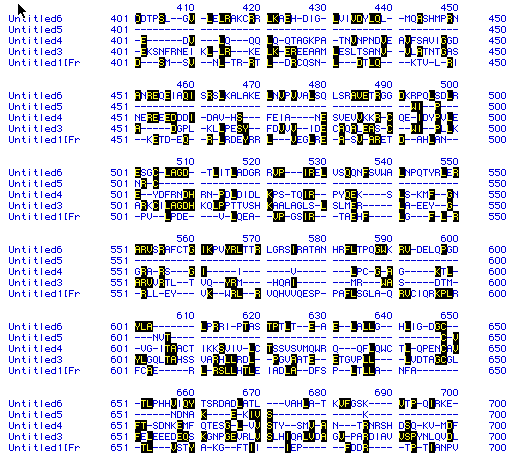 Figure 7. A sample of the results of a multiple sequence alignment
of DNA helicase found in humans,
mice,
yeast,
malarial
parasite, and bacteria.
In the figure, Untitled 6 is from bacteria, 5 is from malarial parastie,
4 is from yeast, 3 is from human, and 1 is from mouse. The black
boxes indicate shared amino acids, and, as you can tell from the alignment,
there isn't much similiarity between the amino acid sequences of DNA helicase
in these different organisms.
Figure 7. A sample of the results of a multiple sequence alignment
of DNA helicase found in humans,
mice,
yeast,
malarial
parasite, and bacteria.
In the figure, Untitled 6 is from bacteria, 5 is from malarial parastie,
4 is from yeast, 3 is from human, and 1 is from mouse. The black
boxes indicate shared amino acids, and, as you can tell from the alignment,
there isn't much similiarity between the amino acid sequences of DNA helicase
in these different organisms.
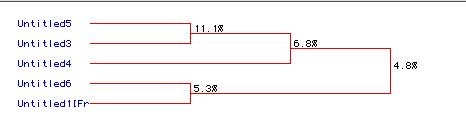 Figure 8. Phylogentic tree of DNA helicase constructed
using degree of amino acid conservation over time. As you can tell, there
is not a high degree of amino acid conservation over time in DNA helicase
in these five organisms, where untitled 6 is bacteria,
5 is malarial
parasite, 4 is yeast,
3 is human,
and 1 is mouse.
The highest degree of conservation is 11.1 % between malarial parasite
and human DNA helicase. The overall conservation is 4.8 %.
Figure 8. Phylogentic tree of DNA helicase constructed
using degree of amino acid conservation over time. As you can tell, there
is not a high degree of amino acid conservation over time in DNA helicase
in these five organisms, where untitled 6 is bacteria,
5 is malarial
parasite, 4 is yeast,
3 is human,
and 1 is mouse.
The highest degree of conservation is 11.1 % between malarial parasite
and human DNA helicase. The overall conservation is 4.8 %.

To return to the DNA helicase page, click here.
To return to the main page, click here.
Email me: schutchins@davidson.edu







